Manolis Kellis - Courses Taught
I teach the following courses on Computational Biology and Algorithms at MIT.
 |
6.881 - Computational Personal Genomics: Making sense of complete genomesWith the growing availability and lowering costs of genotyping and personal genome sequencing, the focus has shifted from the ability to obtain the sequence to the ability to make sense of the resulting information. This course is aimed at exploring the computational challenges associated with interpreting how sequence differences between individuals lead to phenotypic differences such as gene expression, disease predisposition, or response to treatment. It will cover the computational challenges associated with personal genomics, such as genotype phasing and haplotype reconstruction, exploiting linkage for variant imputation, ancestry painting for admixed genomes, predicting likely causal variants using functional or comparative genomics annotations of coding and non-coding elements, relating regulatory variation to gene expression or chromatin state changes, measuring recent evolution and human selection, using systems biology and network information for understanding weak contributions, and the challenge of deciphering complex multi-genic traits such as height, Alzheimer's, diabetes, or cancer. Students will read and discuss seminal papers in the area, download and use existing tools, write their own software to interpret existing human genomes, and discuss limitations, challenges, and ethical, legal and social implications. Previous offerings:
|
 |
6.390 - Intro to Machine LearningLast Taught: Spring 2024 |
 |
6.802/6.874/20.390/20.490 - Computational Systems Biology: Deep Learning in the Life SciencesPresents innovative approaches to computational problems the life sciences focusing on deep learning based approaches with comparisons to conventional methods. |

|
6.7930[6.871]/HST.956 - Machine Learning for Healthcarewith Peter Szolovits.Introduces students to machine learning in healthcare, including the nature of clinical data and the use of machine learning for risk stratification, disease progression modeling, precision medicine, diagnosis, subtype discovery, and improving clinical workflows. Topics include causality, interpretability, algorithmic fairness, time-series analysis, graphical models, deep learning and transfer learning. Guest lectures by clinicians from the Boston area and course projects with real clinical data emphasize subtleties of working with clinical data and translating machine learning into clinical practice. Last taught: Spring 2023 and Fall 2022 |
  |
6.047/6.878 - Computational Biology: Genomes, Networks, Evolutionwith Piotr Indyk (F05, F06), James Galagan (F07, F08, F09), sole in charge (F10, F11).Covers the algorithmic and machine learning foundations of computational biology, combining theory with practice. We study the principles of algorithm design for biological datasets, and analyze influential problems and techniques. We use these to analyze real datasets from large-scale studies in genomics and proteomics. Previous offerings: |
 |
6.046 classic: Introduction to Algorithmswith Ron Rivest (S06), Srini Devadas (S07). Introduction to design and analysis of algorithms. Topics include sorting; search trees, heaps, and hashing; divide-and-conquer; dynamic programming; greedy algorithms; amortized analysis; graph algorithms; shortest paths; network flow. Advanced topics include: number theory, computational biology, string matching, database search, hidden Markov models; number-theoretic algorithms; polynomial and matrix calculations. Previous Offerings: |
 |
6.046 new: Design and Analysis of Algorithmswith Piotr Indyk (S08), Marten van Dijk (S09), Ran Libeskind-Hadas (S14), and Mikhail Kapralov (S14). With the creation of 6.006, the new 6.046 became a more advanced algorithms course in 2008 (using 6.006 as a pre-requisite), mostly serving as an entry point to theory courses, and targeting junior and senior students interested in specializing in theory. Previous Offerings: |
 |
6.006: Introduction to Algorithms (new)with Piotr Indyk and Erik Demaine (S11) In contrast, 6.006 became a more practical course with programming assignments and a greater emphasis on real-world applications of the algorithms and techniques taught. |
 |
6.092: Bioinformatics and Proteomics: An Engineering-Based Problem Solving ApproachGil Alterovitz, Manolis Kellis, Marco Ramoni This interdisciplinary course provides a hands-on approach to students in the topics of bioinformatics and proteomics. Lectures and labs cover sequence analysis, microarray expression analysis, Bayesian methods, control theory, scale-free networks, and biotechnology applications. Designed for those with a computational and/or engineering background, it will include current real-world examples, actual implementations, and engineering design issues. Where applicable, engineering issues from signal processing, network theory, machine learning, robotics and other domains will be expounded upon. New research areas will be explored using current literature as well as text from book chapter materials being written by the instructors. Guest lectures include speakers from both industry and academia. (IAP 2005) |
    |
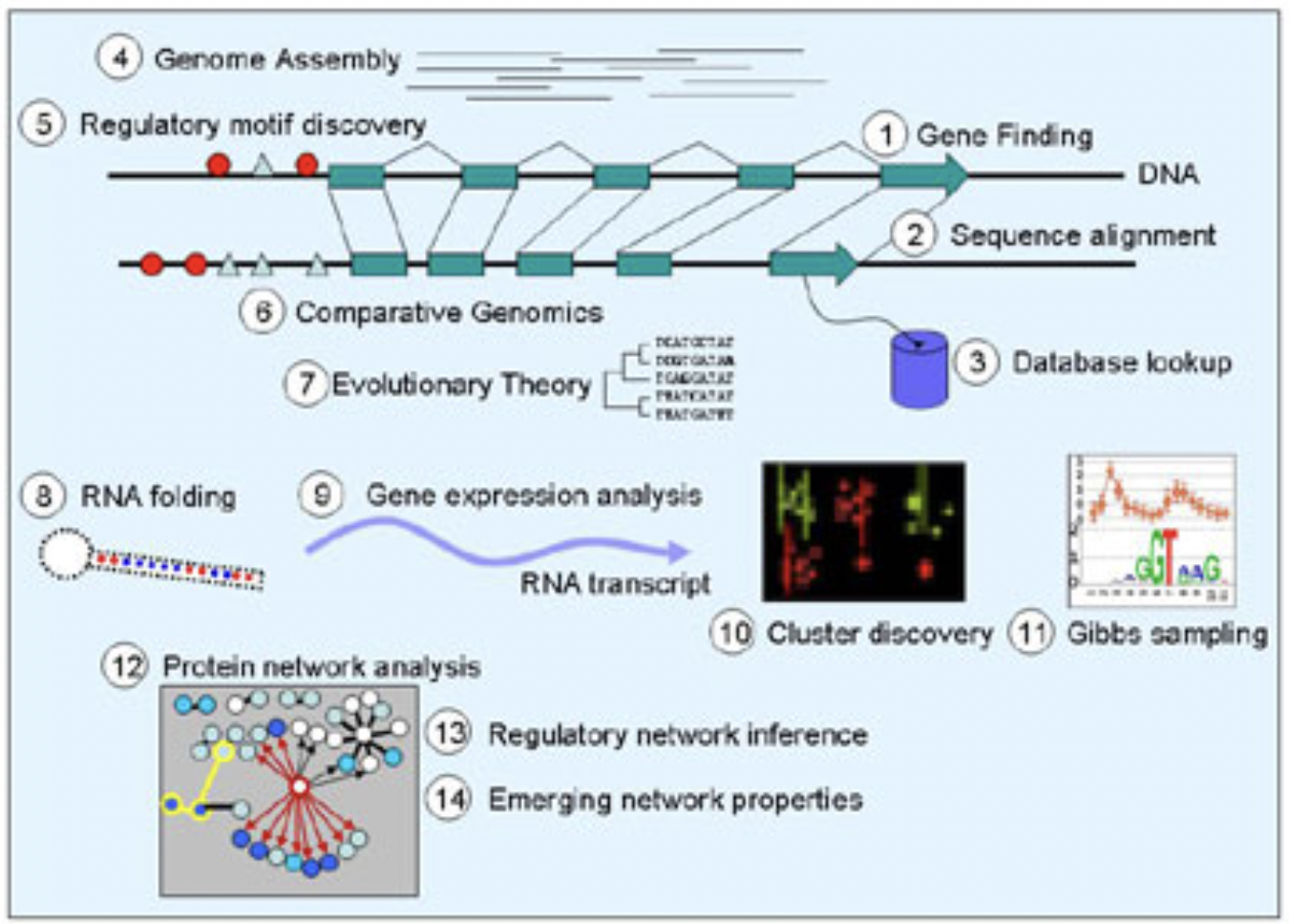
6.096 - Algorithms for Computational Biology (meets with 6.046)Manolis KellisThis course covers the algorithmic foundations of computational biology, combining theory with practice. We study the principles of algorithm design for biological datasets, analyze influential algorithms, and apply these to real datasets. Topics include: biological sequence analysis, gene finding, motif discovery, RNA folding, global and local sequence alignment, genome assembly, comparative genomics, genome duplication, genome rearrangements, evolutionary theory, gene expression, clustering algorithms, scale-free networks, machine learning applications to genomics. (Spring 2005) |
Other Computational Biology Courses offered at MIT
| Course | Title | Professor |
|---|---|---|
| 6.047/6.878 | Computational Biology: Genomes, Networks, Evolution | M. Kellis (with P. Indyk and J. Galagan) |
| 6.891 | Computational Evolutionary Biology | R. Berwick |
| 6.892/7.90 | Computational Functional Genomics | D. Gifford, T. Jaakkola, R. Young |
| 7.81J/8.591 | Systems Biology | A. vanOudenaarden |
| 7.91 | Foundations of Computational and Systems Biology | M. Yaffe, C. Burge, A. Keating |
| 10.555 | BioInformatics | I. Rigoutsos, G. Stephanopoulos |
| 18.417 | Introduction to Computational Molecular Biology | J. Waldispuhl, B. Berger |
| HST.508 | Quantitative Genomics and Evolution | L. Mirny |
| 6.096 | Algorithms for Computational Biology | M. Kellis |
