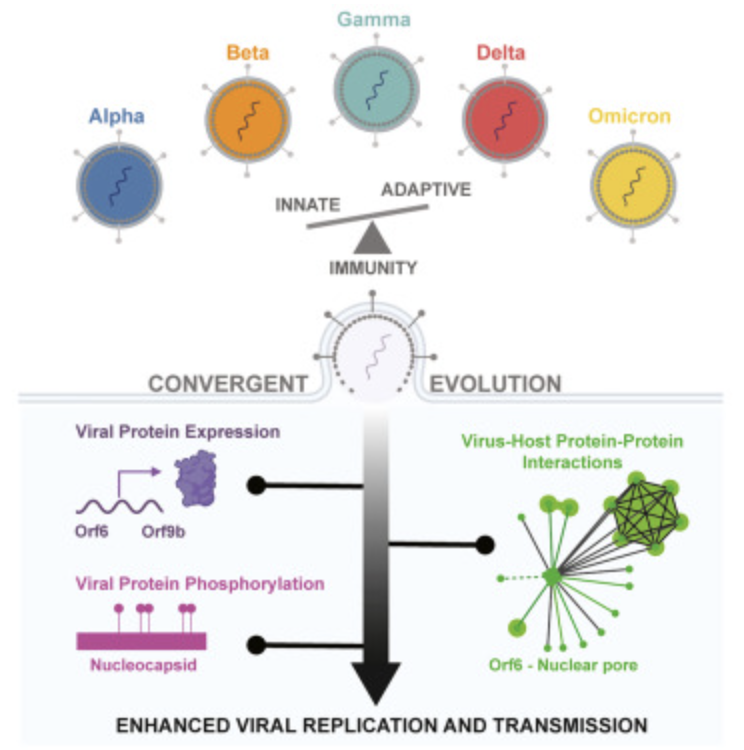
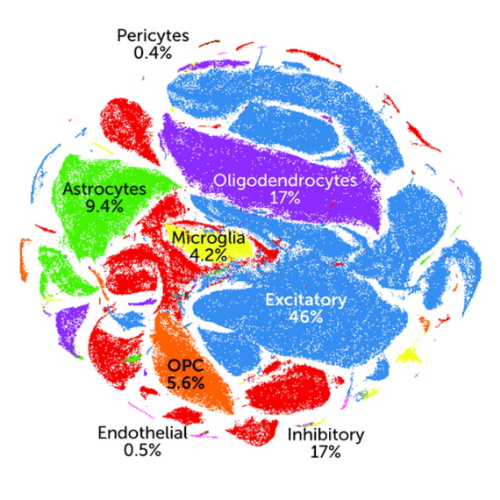
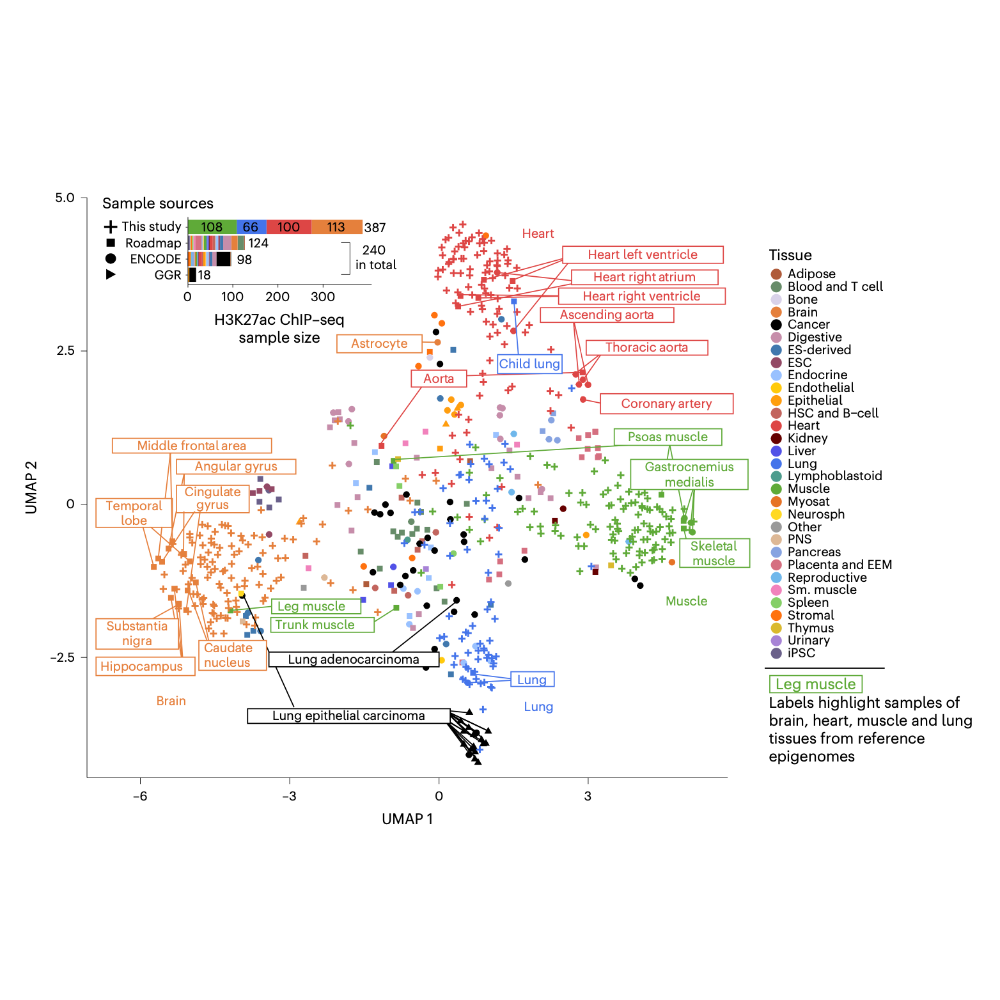
Mission
We seek to understand the mechanistic basis of human disease using a combination of computational and experimental techniques.
This involves developing methods for:
- the systematic discovery and characterization of functional elements in the human genome
- the discovery and validation of the gene-regulatory circuitry controlling these elements
- the use of epigenomic information for annotating regulatory regions and their activity across different cell types
- the use of comparative genomics for recognizing coding and non-coding regions of functional importance for evolutionary fitness
We work in a highly interdisciplinary environment at the interface of computer science and biology. Members of the group come from a primarily computational background and share a strong passion for understanding biological systems. We are engaged in several collaborative research partnerships with biological and experimental collaborators, at the Broad Institute, the ENCODE, modENCODE, GTEx, and Epigenomics Roadmap consortia, the Harvard Medical School, and other universities.