Ongoing Projects
| See a static view grouped by research area: |
Single-cell Genomics
- Variation and Disease
- Genome Interpretation
- Brain Genomics/Alzheimer's/Schizophrenia/Neuroscience
- Metabolism/Obesity/Diabetes/Exercise
- Cancer Genomics
- RNA Biology
- Small RNAs
- Genes
- Epigenomics
- Regulatory Motifs
- Bio Networks
- Evolution
- Phylogenomics
- Algorithms
- Geometry/Vision
- Robotics/AI
- Editorials. |
See a dynamic list of our papers, select multiple areas, and search authors or keywords: Paper Explorer browser.
Single-Cell Genomics
Dissecting disease circuitry requires understanding the specific cell type of action where genetic variants exert their effects, in order to guide the development of new therapeutics and of the right assays for understanding disease mechanism. These cell types can often be hidden as only a small fraction of complex tissues, and thus can be missed when profiling bulk tissue. A major effort in our group to overcome these challenges is to develop and systematically apply single-cell transcriptomic and epigenomic profiling in complex tissue samples across hundreds of individuals, to recognize the cell type of action of major disorders, and the genes and regulatory regions whose activity is altered in each of the cell types.
 |
260.
A multiresolution framework to characterize single-cell state landscapes. (pdf)
Journal: Nature Communications, 2020 Oct 26 doi: 10.1038/s41467-020-18416-6 PMID:33106496 PMCID: PMC7588427 |
 |
212.
Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. (pdf)
Journal: Cell Reports, 2019 Dec 24 doi: 10.1016/j.celrep.2019.11.083 PMID:31875561 PMCID: PMC6939677 |
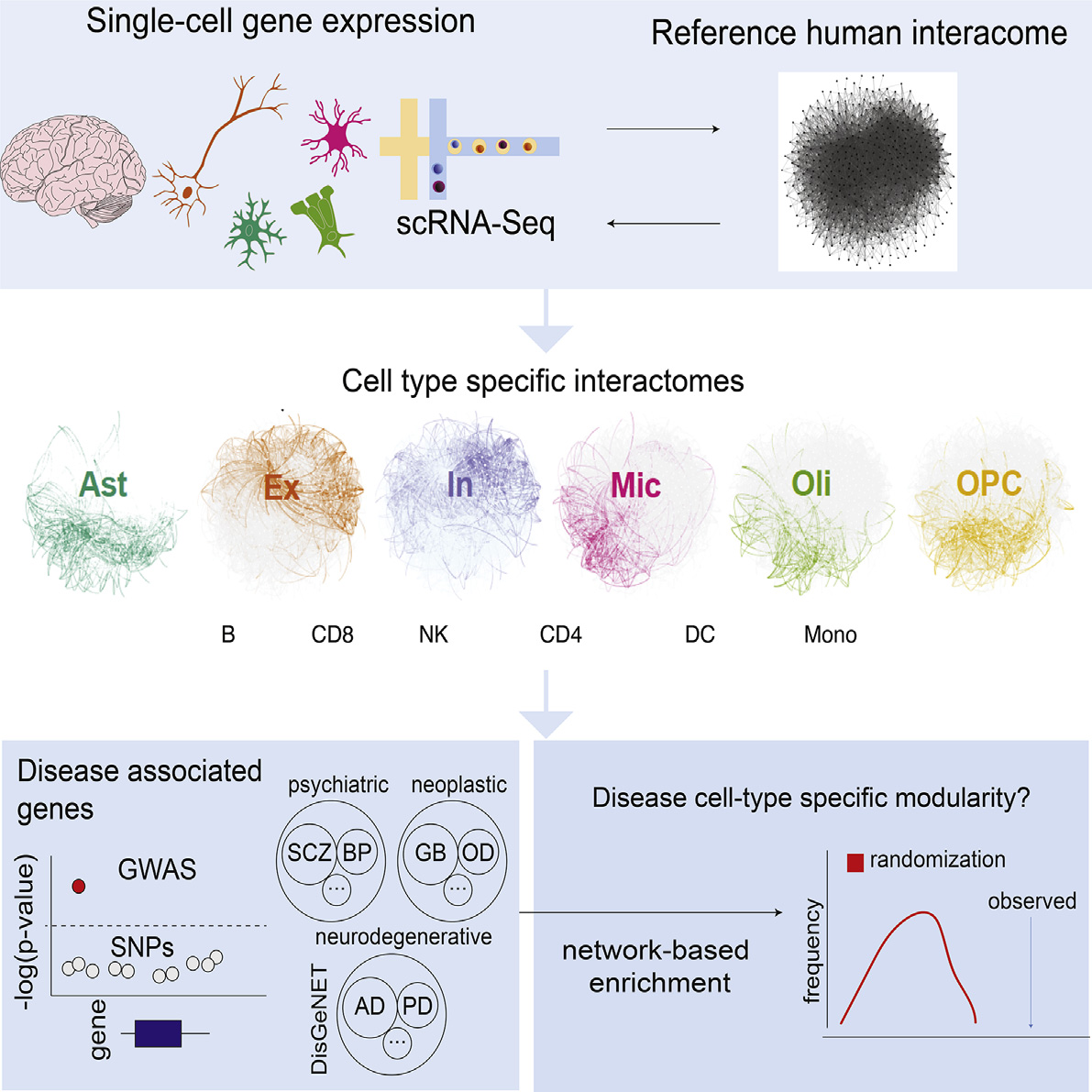 |
211.
Reconstruction of Cell-type-Specific Interactomes at Single-Cell Resolution. (pdf)
Journal: Cell Systems, 2019 Dec 18 doi: 10.1016/j.cels.2019.10.007 PMID:31786210 PMCID: PMC6943823 |
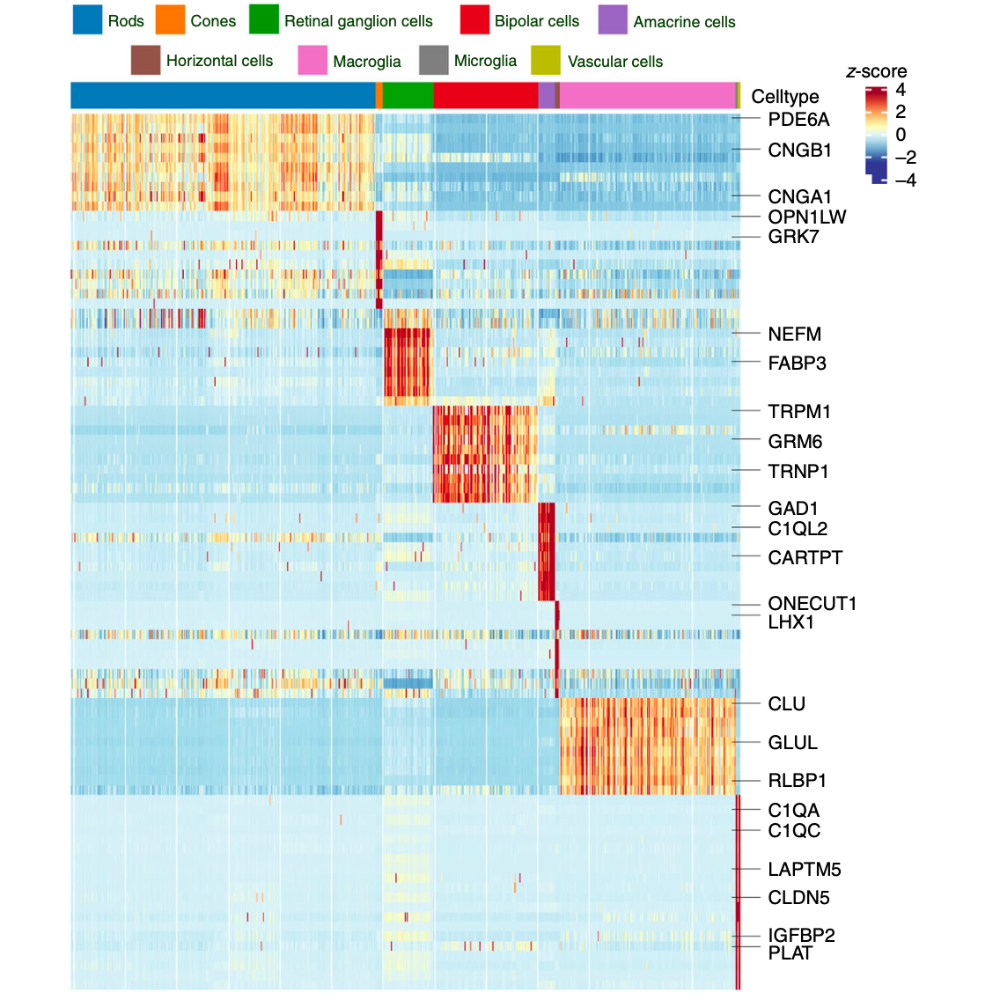 |
209.
Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. (pdf)
Journal: Nature Communications, 2019 Oct 25 doi: 10.1038/s41467-019-12780-8 PMID:31653841 PMCID: PMC6814749 |
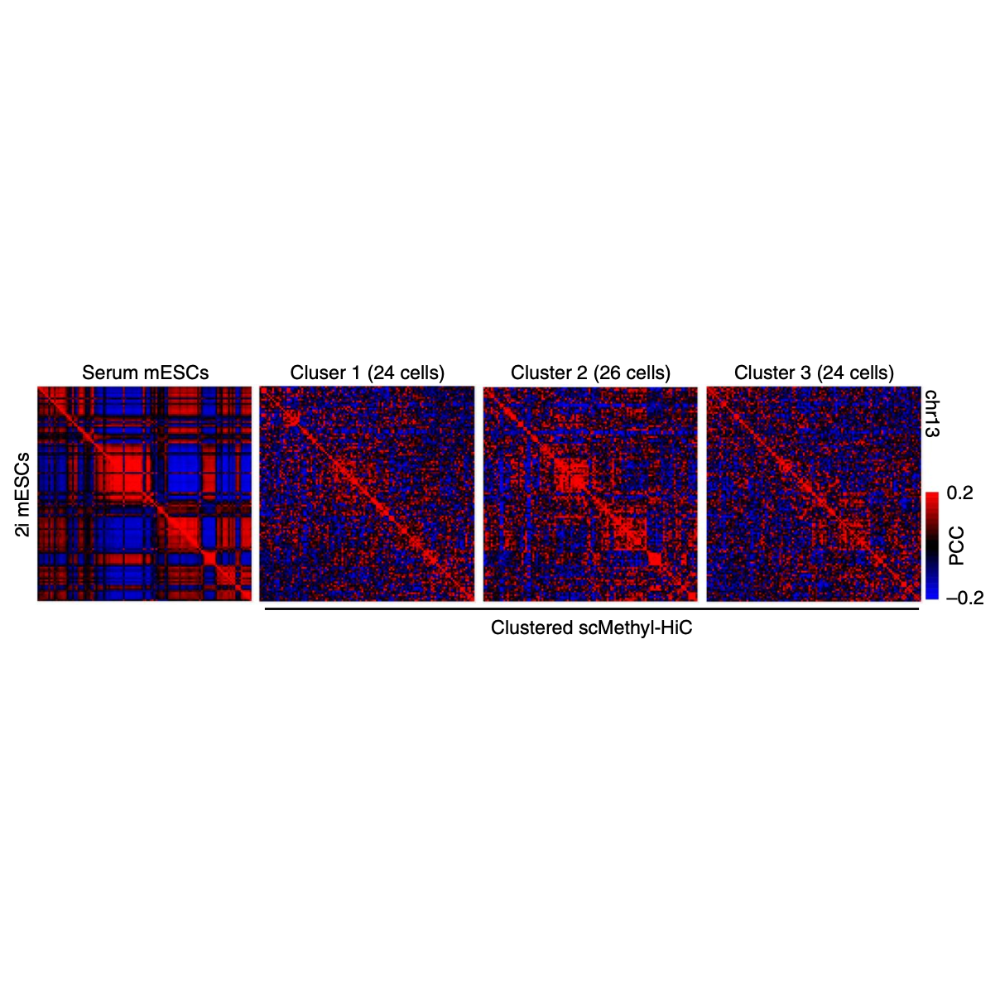 |
207.
Joint profiling of DNA methylation and chromatin architecture in single cells. (pdf)
Journal: Nature Methods, 2019 Oct doi: 10.1038/s41592-019-0502-z PMID:31384045 PMCID: PMC6765429 |
 |
203.
Single-cell transcriptomic analysis of Alzheimer's disease. (pdf)
Journal: Nature, 2019 Jun doi: 10.1038/s41586-019-1195-2 PMID:31042697 PMCID: PMC6865822 |
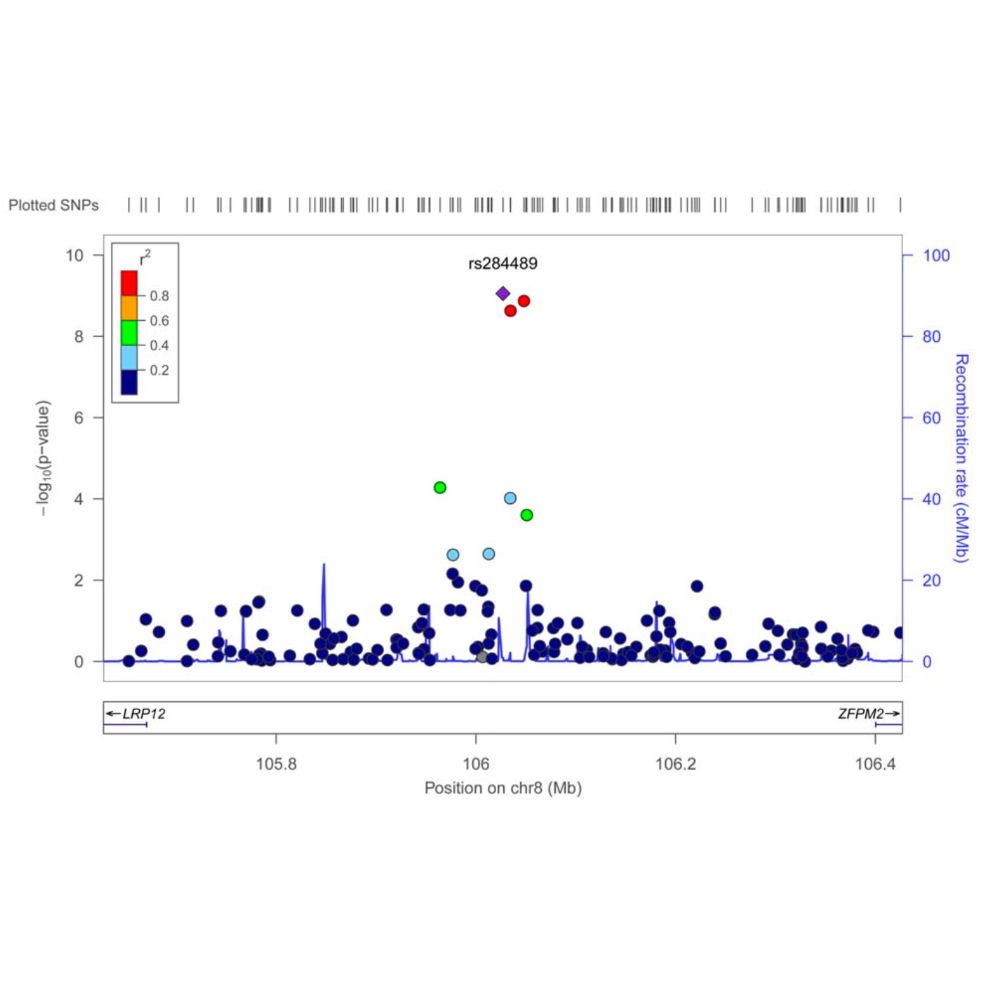
Variation and Disease
A major focus of our lab is understanding the effects of genetic variation on molecular phenotypes and human disease. We develop methods for integrating diverse functional genomic datasets of transcription, chromatin modifications, regulator binding, and their changes across multiple conditions to interpret genetic associations, identify causal variants, and predict the effects of genetic perturbations.
 |
270.
Distinct metabolic programs established in the thymus control effector functions of gammadelta T cell subsets in tumor microenvironments. (pdf)
Journal: Nature Immunology, 2021 Feb doi: 10.1038/s41590-020-00848-3 PMID:33462452 PMCID: PMC7610600 |
 |
268.
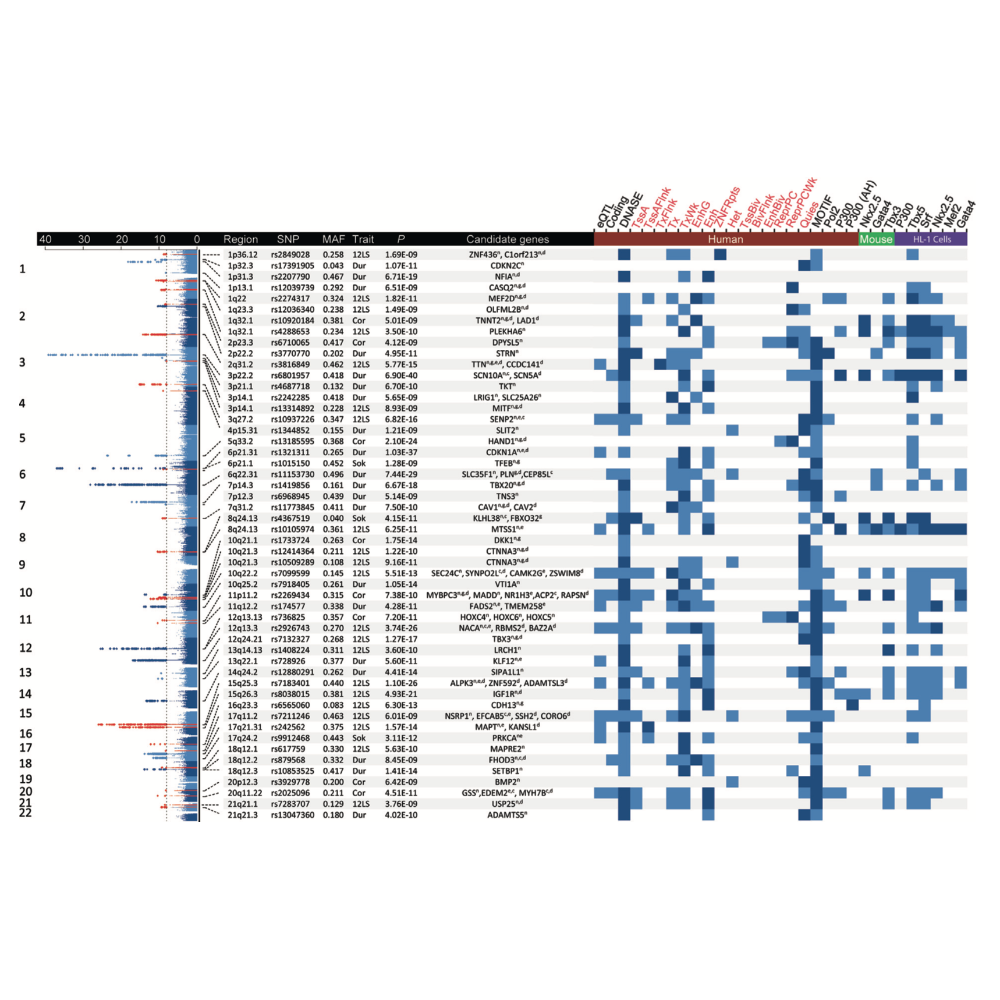
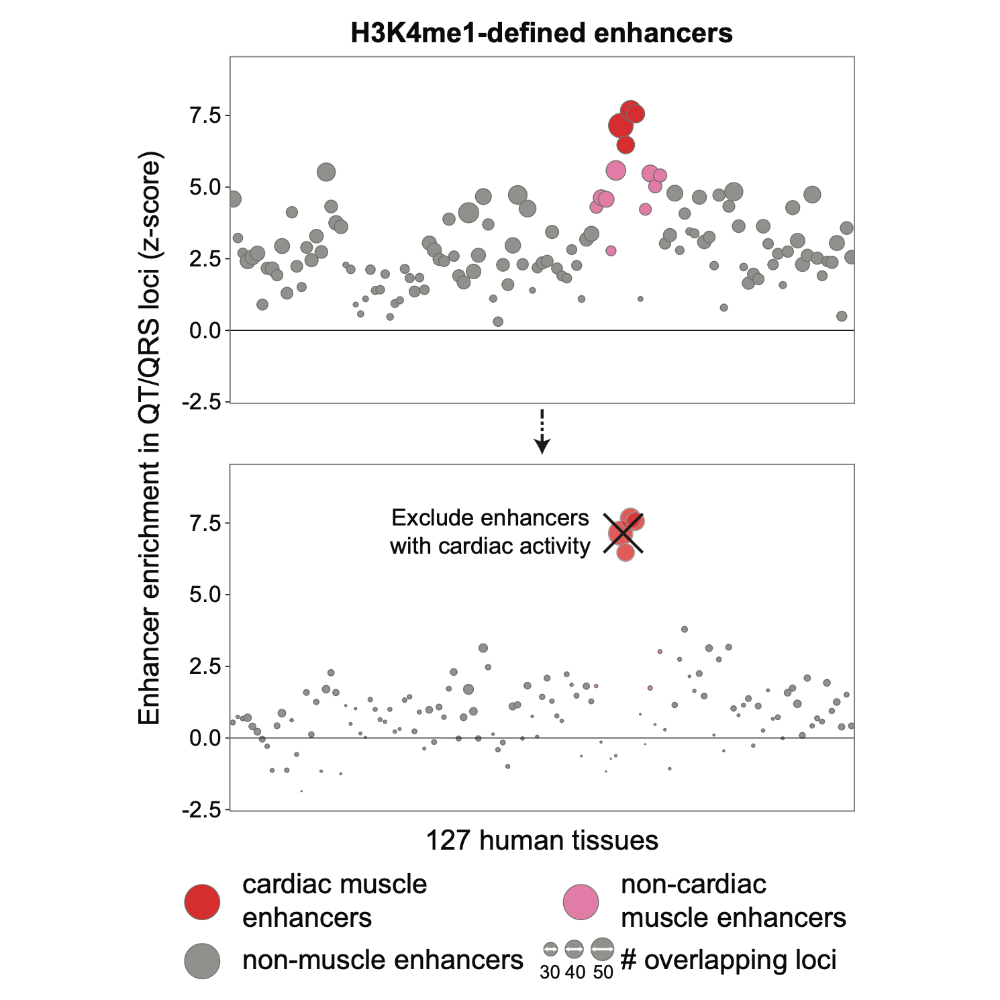
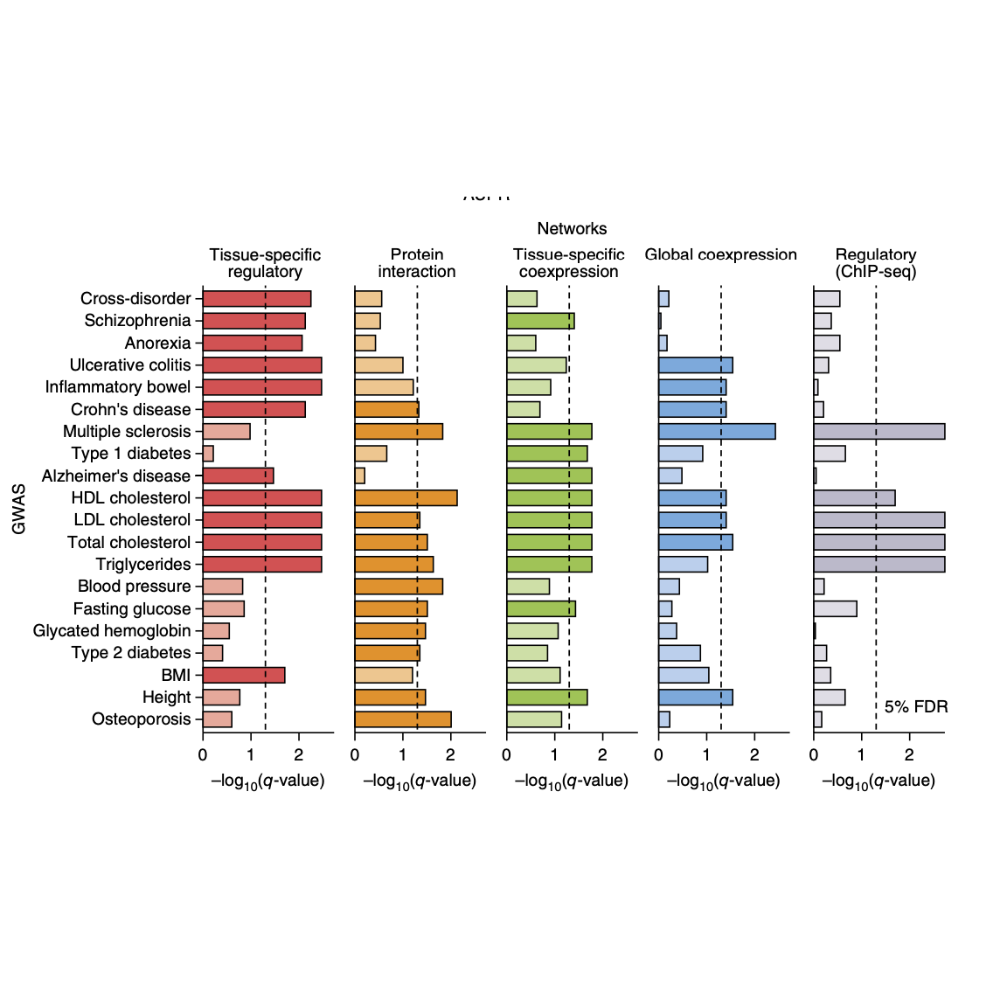
Regulatory genomic circuitry of human disease loci by integrative epigenomics. (pdf)
Journal: Nature, 2021 Feb doi: 10.1038/s41586-020-03145-z PMID:33536621 PMCID: PMC7875769 |
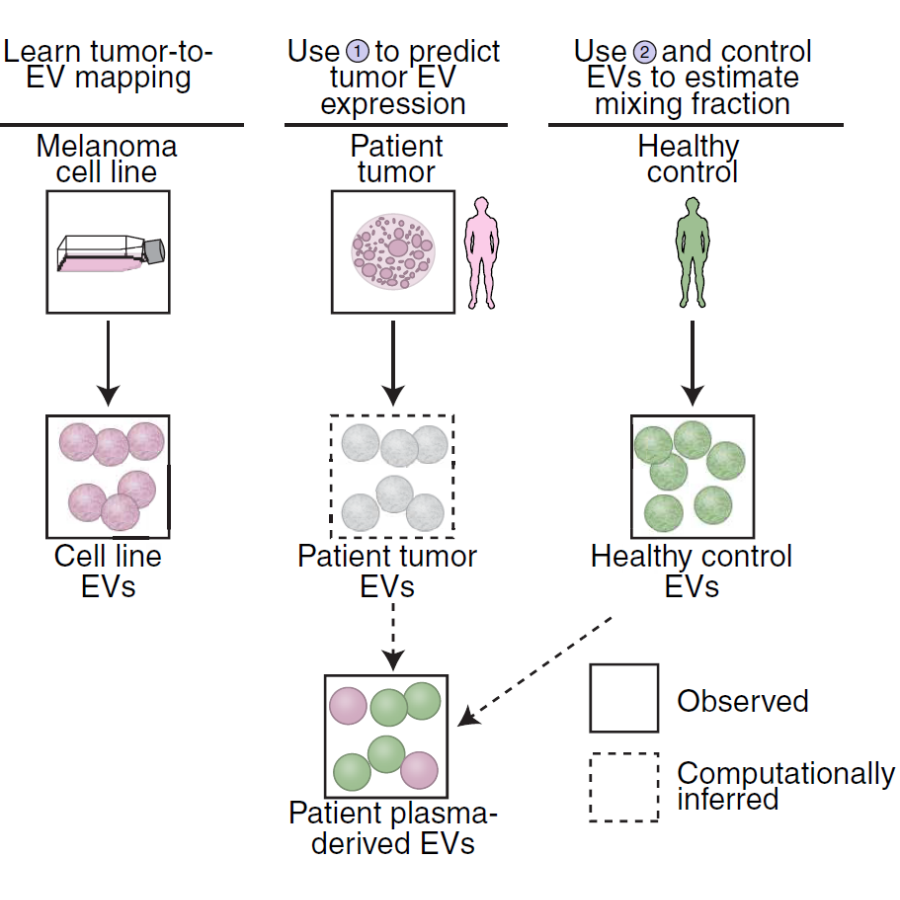 |
262.
Plasma-derived extracellular vesicle analysis and deconvolution enable prediction and tracking of melanoma checkpoint blockade outcome. (pdf)
Journal: Science Advances, 2020 Nov doi: 10.1126/sciadv.abb3461 PMID:33188016 PMCID: PMC7673759 |
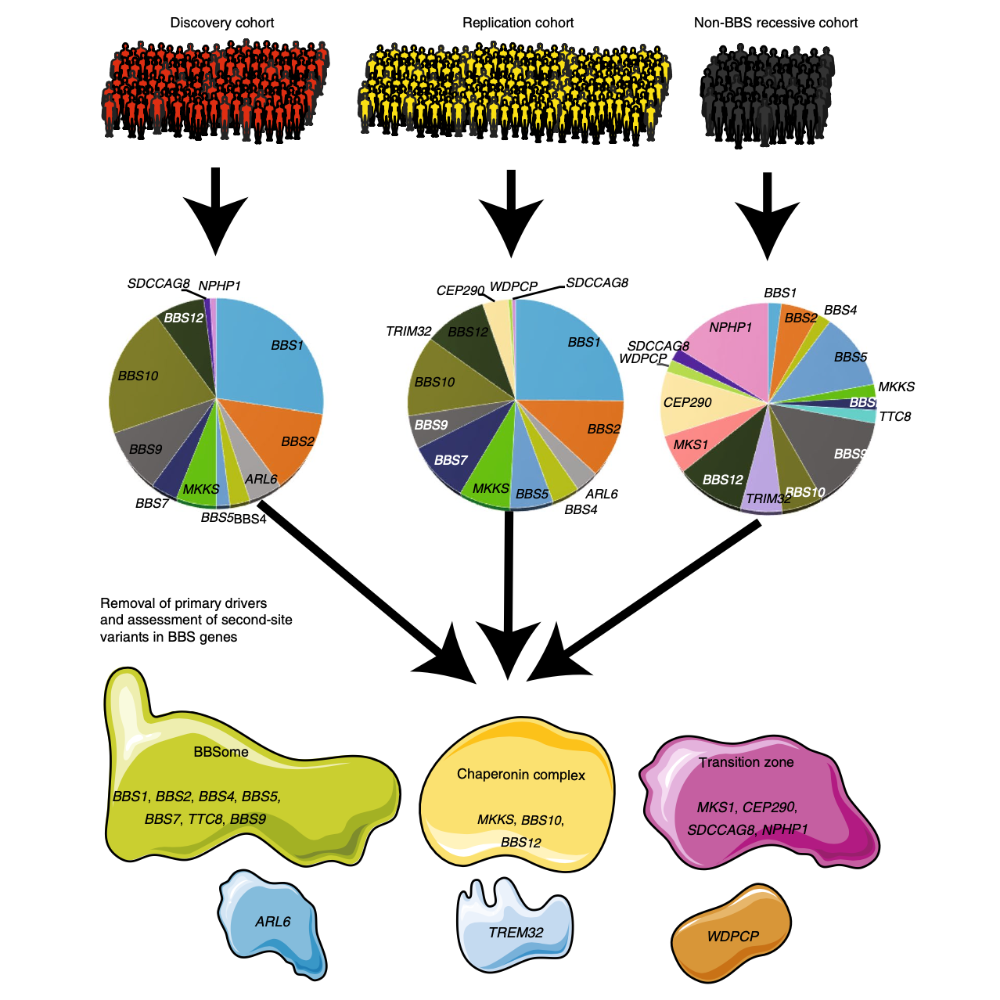 |
261.
Evidence for secondary-variant genetic burden and non-random distribution across biological modules in a recessive ciliopathy. (pdf)
Journal: Nature Genetics, 2020 Nov doi: 10.1038/s41588-020-0707-1 PMID:33046855 PMCID: PMC8272915 |
 |
256.
Improved haplotype inference by exploiting long-range linking and allelic imbalance in RNA-seq datasets. (pdf)
Journal: Nature Communications, 2020 Sep 16 doi: 10.1038/s41467-020-18320-z PMID:32938926 PMCID: PMC7494856 |
 |
252.
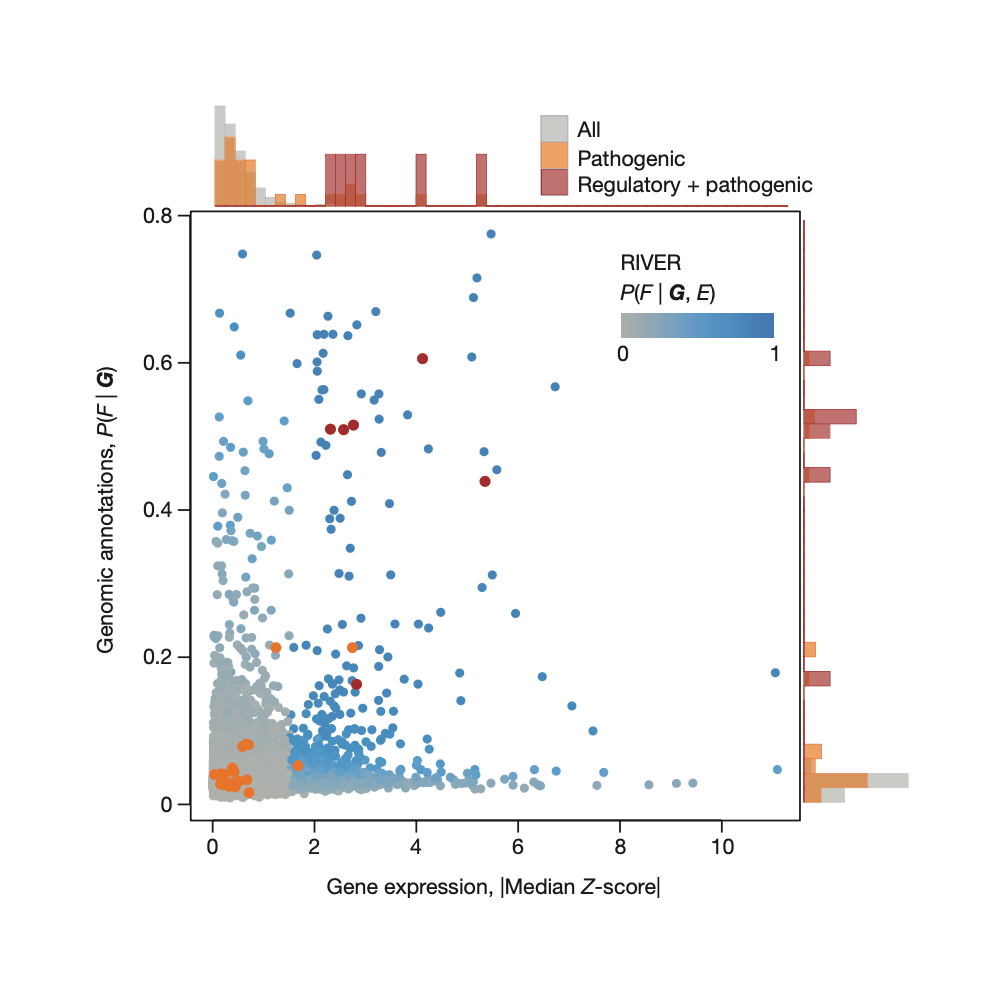
Transcriptomic signatures across human tissues identify functional rare genetic variation. (pdf)
Journal: Science, 2020 Sep 11 doi: 10.1126/science.aaz5900 PMID:32913073 PMCID: PMC7646251 |
 |
243.
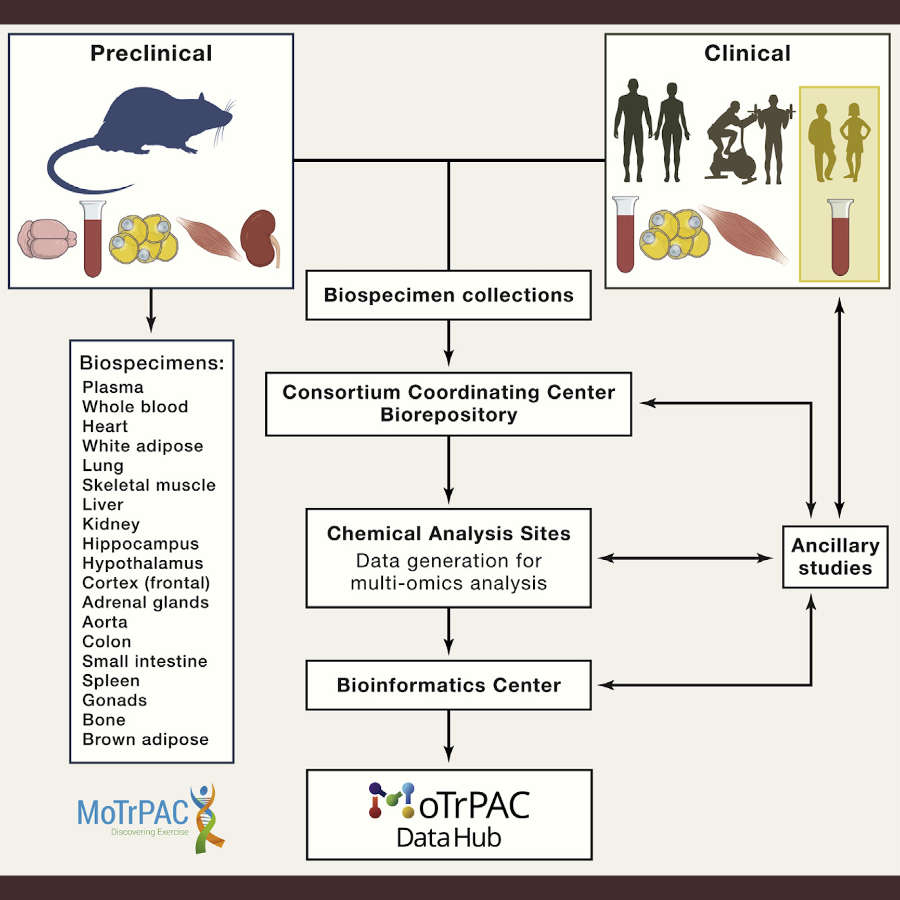
Molecular Transducers of Physical Activity Consortium (MoTrPAC): Mapping the Dynamic Responses to Exercise. (pdf)
Journal: Cell, 2020 Jun 25 doi: 10.1016/j.cell.2020.06.004 PMID:32589957 PMCID: PMC8800485 |
 |
239.
Inferring multimodal latent topics from electronic health records. (pdf)
Journal: Nature Communications, 2020 May 21 doi: 10.1038/s41467-020-16378-3 PMID:32439869 PMCID: PMC7242436 |
 |
237.
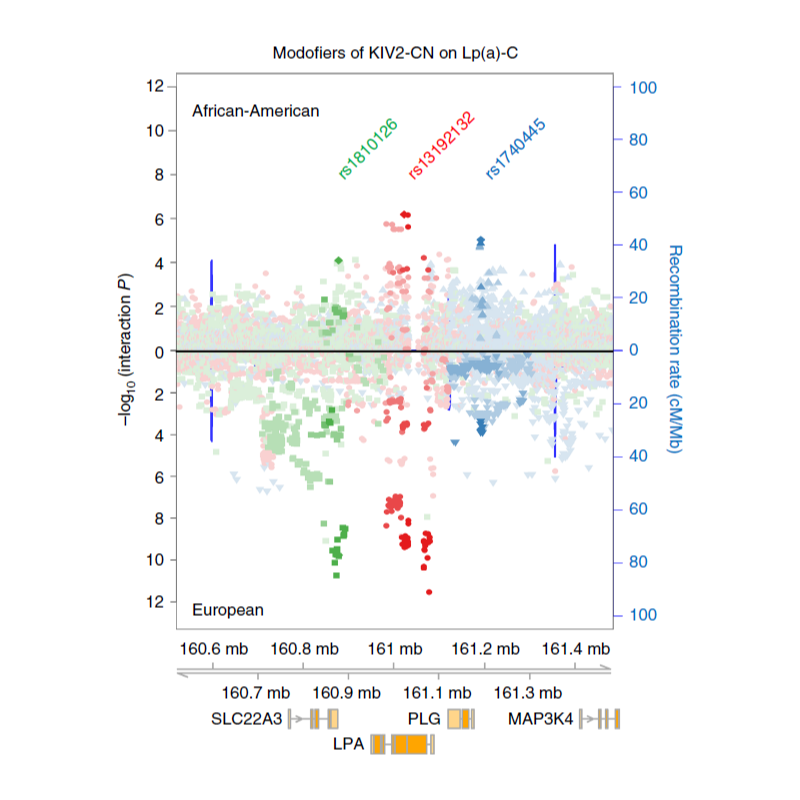
Publisher Correction: Deep coverage whole genome sequences and plasma lipoprotein(a) in individuals of European and African ancestries. (pdf)
Journal: Nature Communications, 2020 Apr 1 doi: 10.1038/s41467-020-15236-6 PMID:32238811 PMCID: PMC7113276 |
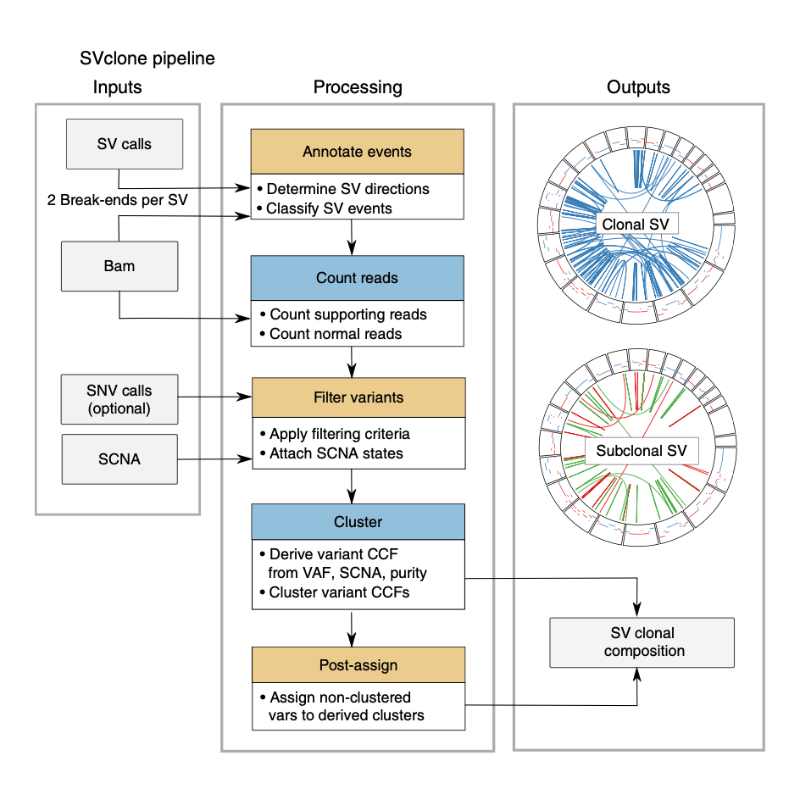 |
219.
Inferring structural variant cancer cell fraction. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-020-14351-8 PMID:32024845 PMCID: PMC7002525 |
 |
216.
Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. (pdf)
Journal: Nature, 2020 Feb doi: 10.1038/s41586-020-1965-x PMID:32025015 PMCID: PMC7054214 |
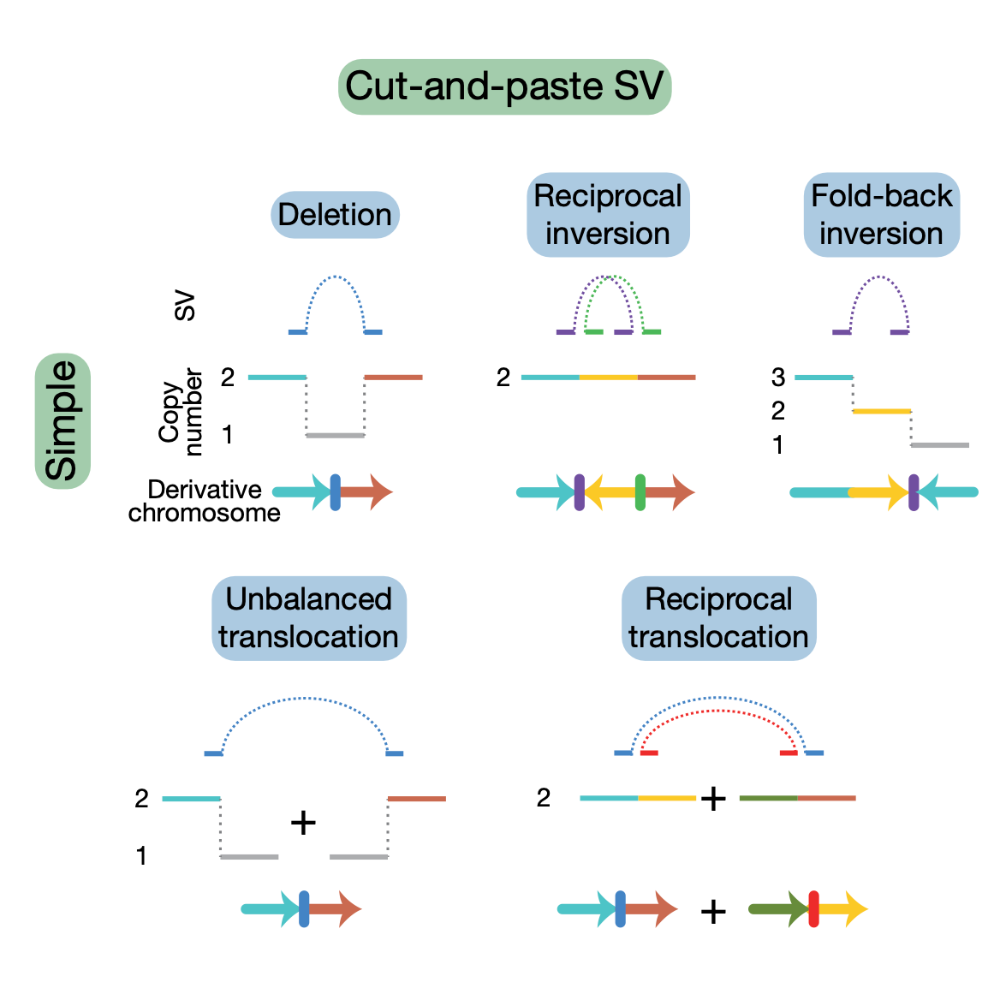 |
215.
Patterns of somatic structural variation in human cancer genomes. (pdf)
Journal: Nature, 2020 Feb doi: 10.1038/s41586-019-1913-9 PMID:32025012 PMCID: PMC7025897 |
 |
204.
Rate of brain aging and APOE epsilon4 are synergistic risk factors for Alzheimer's disease. (pdf)
Journal: Life Science Alliance, 2019 Jun doi: 10.26508/lsa.201900303 PMID:31133613 PMCID: PMC6537750 |
 |
202.
Challenges in IBD Research: Environmental Triggers. (pdf)
Journal: Inflammatory Bowel Diseases, 2019 May 16 doi: 10.1093/ibd/izz076 PMID:31095702 PMCID: PMC6787673 |
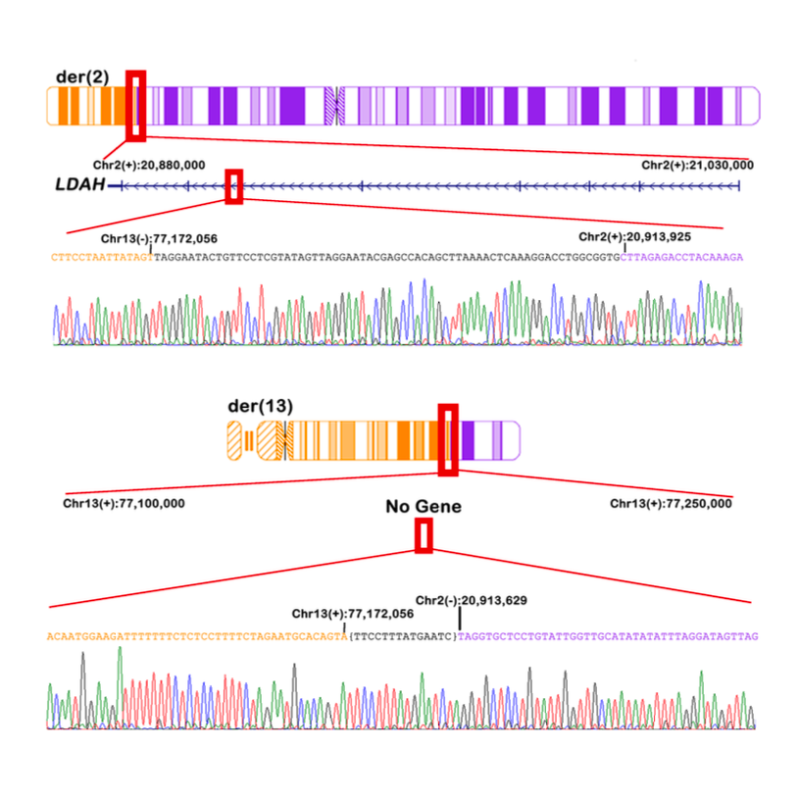 |
201.
Corrigendum: Loss of LDAH associated with prostate cancer and hearing loss. (pdf)
Journal: Human Molecular Genetics, 2019 May 15 doi: 10.1093/hmg/ddz036 PMID:31222336 PMCID: PMC6494788 |
 |
199.
Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. (pdf)
Journal: Nature Genetics, 2019 Feb doi: 10.1038/s41588-018-0307-5 PMID:30643251 PMCID: PMC6358542 |
 |
188.
Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. (pdf)
Journal: Nature Communications, 2018 Aug 23 doi: 10.1038/s41467-018-05747-8 PMID:30140000 PMCID: PMC6107638 |
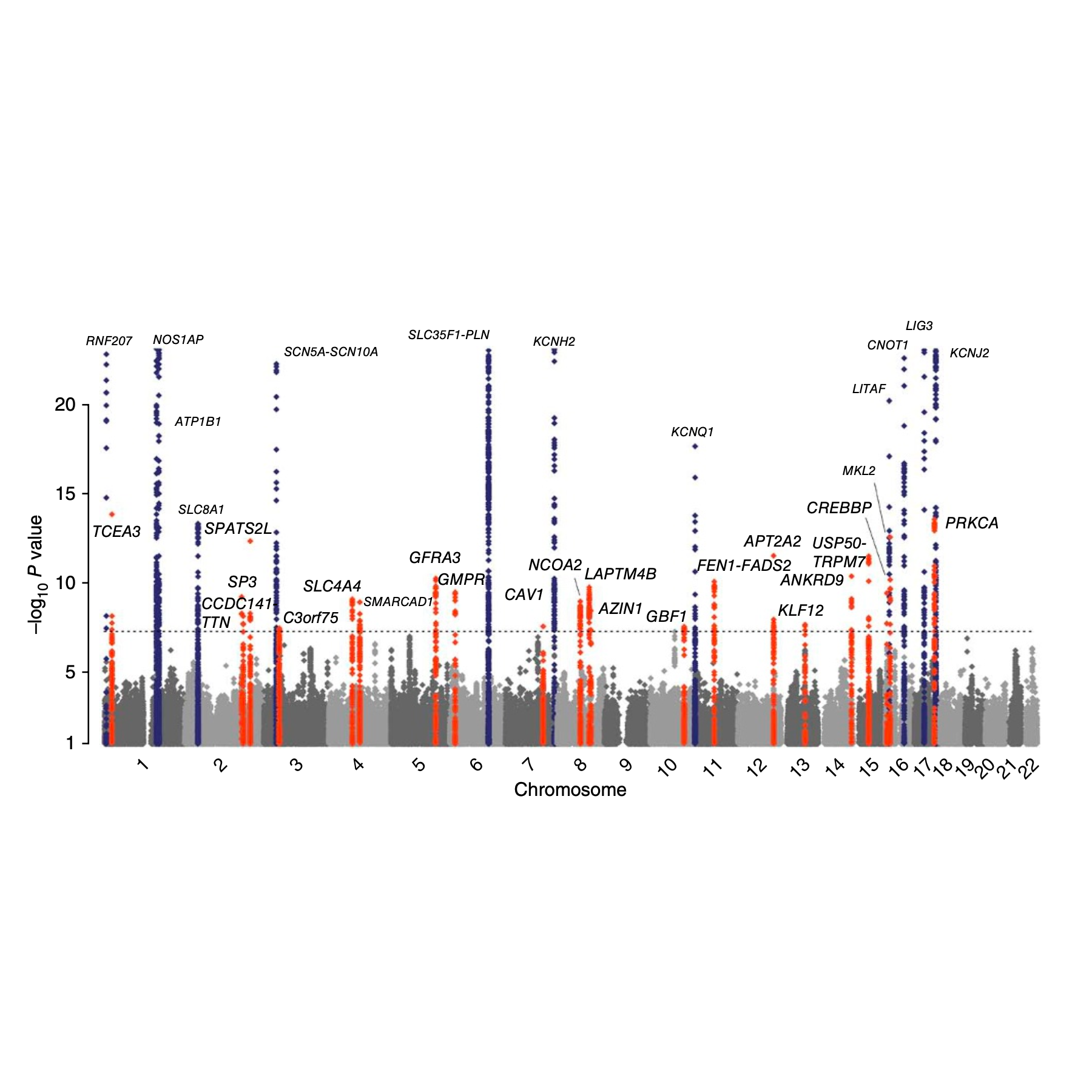
 |
182.
Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. (pdf)
Journal: Nature Genetics, 2018 Jul doi: 10.1038/s41588-018-0154-4 PMID:29955180 PMCID: PMC6248311 |
 |
175.
Identifying cis-mediators for trans-eQTLs across many human tissues using genomic mediation analysis. (pdf)
Journal: Genome Research, 2017 Nov doi: 10.1101/gr.216754.116 PMID:29021290 PMCID: PMC5668943 |
 |
173.
Genetic effects on gene expression across human tissues. (pdf)
Journal: Nature, 2017 Oct 11 doi: 10.1038/nature24277 PMID:29022597 PMCID: PMC5776756 |
 |
171.
The impact of rare variation on gene expression across tissues. (pdf)
Journal: Nature, 2017 Oct 11 doi: 10.1038/nature24267 PMID:29022581 PMCID: PMC5877409 |
 |
165.
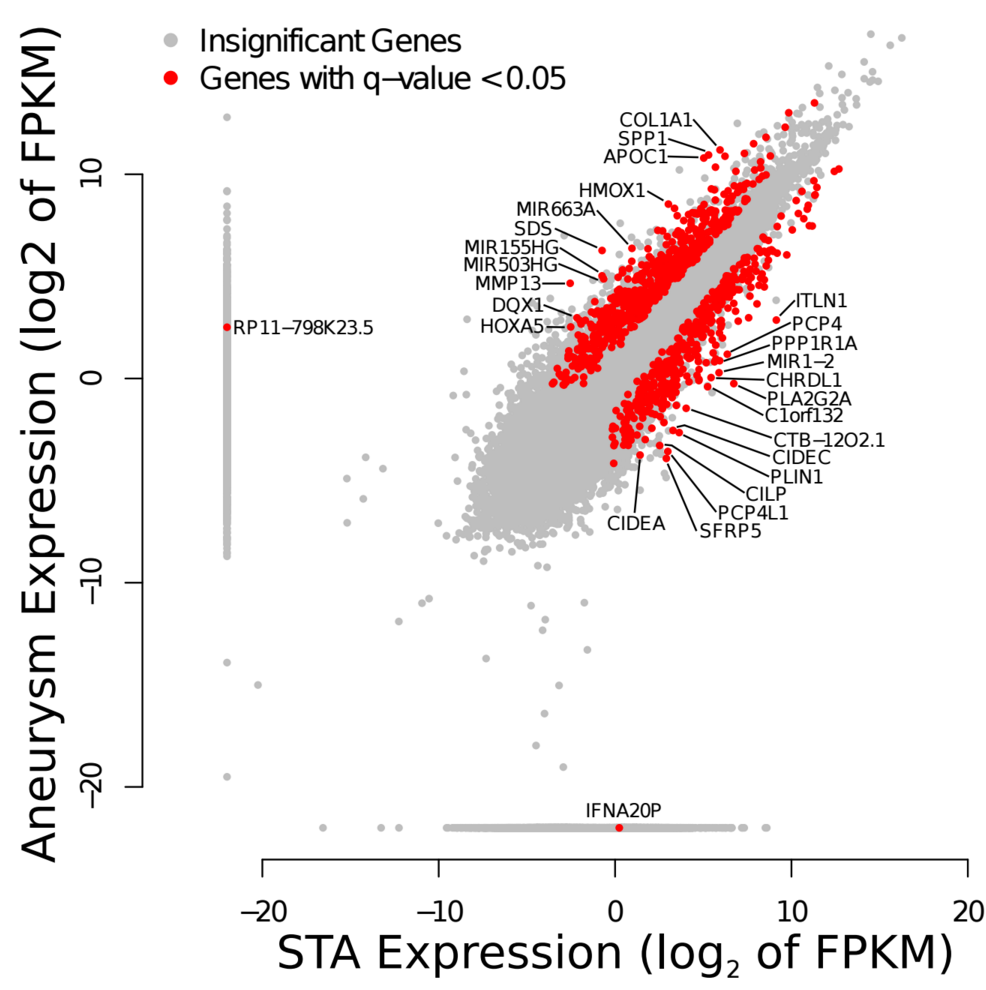
MicroRNA and gene expression changes in unruptured human cerebral aneurysms. (pdf)
Journal: Journal Of Neurosurgery, 2016 Dec doi: 10.3171/2015.11.JNS151841 PMID:26918470 PMCID: PMC5001931 |
 |
161.
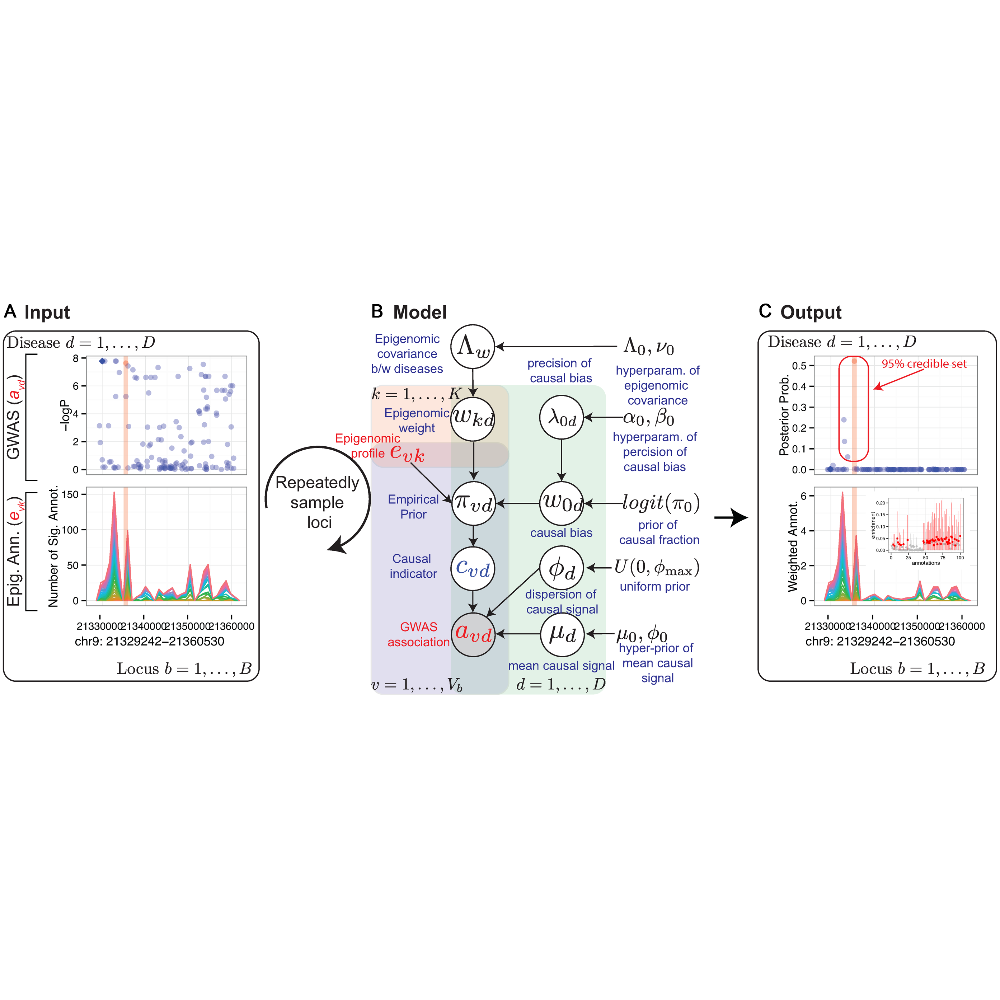
Joint Bayesian inference of risk variants and tissue-specific epigenomic enrichments across multiple complex human diseases. (pdf)
Journal: Nucleic Acids Research, 2016 Oct 14 doi: 10.1093/nar/gkw627 PMID:27407109 PMCID: PMC5062982 |
 |
160.
52 Genetic Loci Influencing Myocardial Mass. (pdf)
Journal: Journal Of The American College Of Cardiology, 2016 Sep 27 doi: 10.1016/j.jacc.2016.07.729 PMID:27659466 PMCID: PMC5478167 |
 |
159.
Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. (pdf)
Journal: Elife, 2016 May 10 doi: 10.7554/eLife.10557 PMID:27162171 PMCID: PMC4862755 |
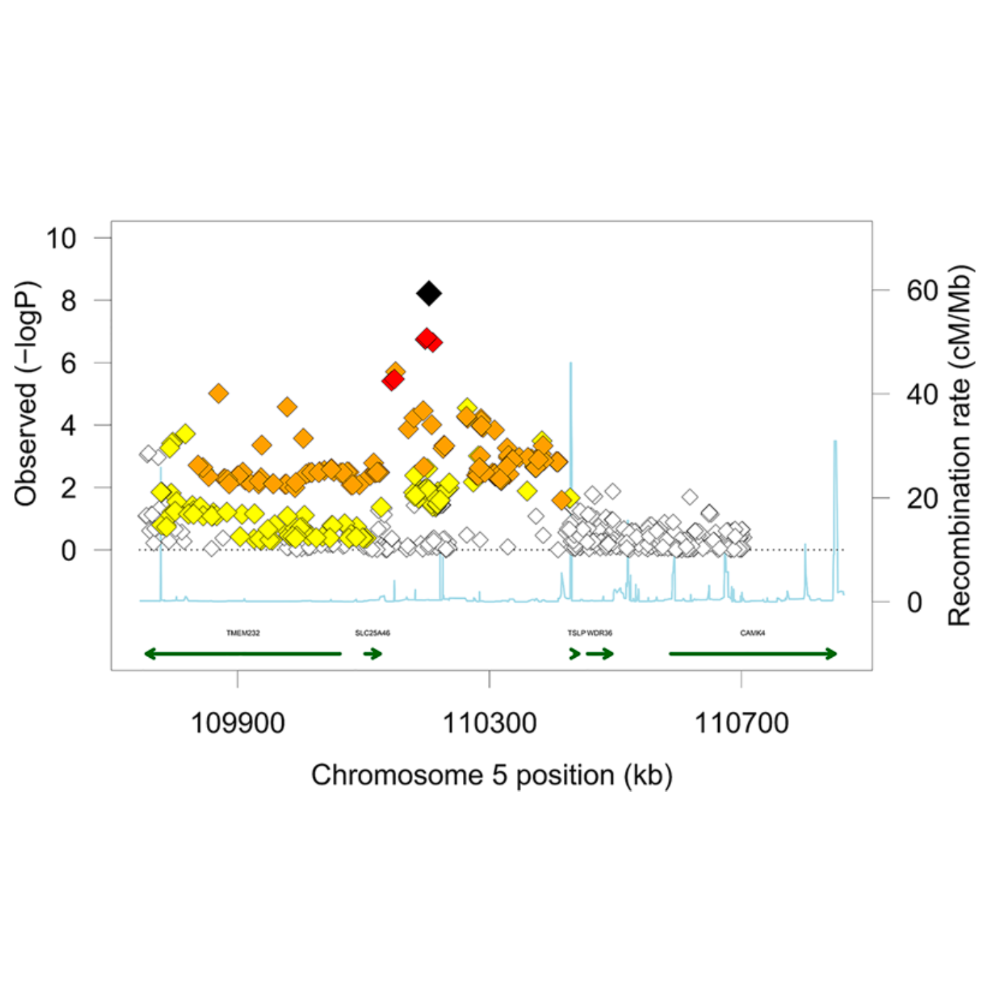 |
158.
Discovery of Genetic Variation on Chromosome 5q22 Associated with Mortality in Heart Failure. (pdf)
Journal: Plos Genetics, 2016 May doi: 10.1371/journal.pgen.1006034 PMID:27149122 PMCID: PMC4858216 |
 |
156.
Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. (pdf)
Journal: Nature Methods, 2016 Apr doi: 10.1038/nmeth.3799 PMID:26950747 PMCID: PMC4967716 |
 |
155.
Survey of variation in human transcription factors reveals prevalent DNA binding changes. (pdf)
Journal: Science, 2016 Mar 25 doi: 10.1126/science.aad2257 PMID:27013732 PMCID: PMC4825693 |
 |
154.
Corrigendum: Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. (pdf)
Journal: Scientific Reports, 2016 Jan 21 doi: 10.1038/srep19384 PMID:26795431 PMCID: PMC4726320 |
 |
153.
FTO Obesity Variant and Adipocyte Browning in Humans. (pdf)
Journal: The New England Journal Of Medicine, 2016 Jan 14 doi: 10.1056/NEJMc1513316 PMID:26760096 |
 |
152.
HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. (pdf)
Journal: Nucleic Acids Research, 2016 Jan 4 doi: 10.1093/nar/gkv1340 PMID:26657631 PMCID: PMC4702929 |
 |
151.
Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. (pdf)
Journal: Scientific Reports, 2015 Oct 19 doi: 10.1038/srep15145 PMID:26477495 PMCID: PMC4609956 |
 |
150.
A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. (pdf)
Journal: Biopreservation And Biobanking, 2015 Oct doi: 10.1089/bio.2015.0032 PMID:26484571 PMCID: PMC4675181 |
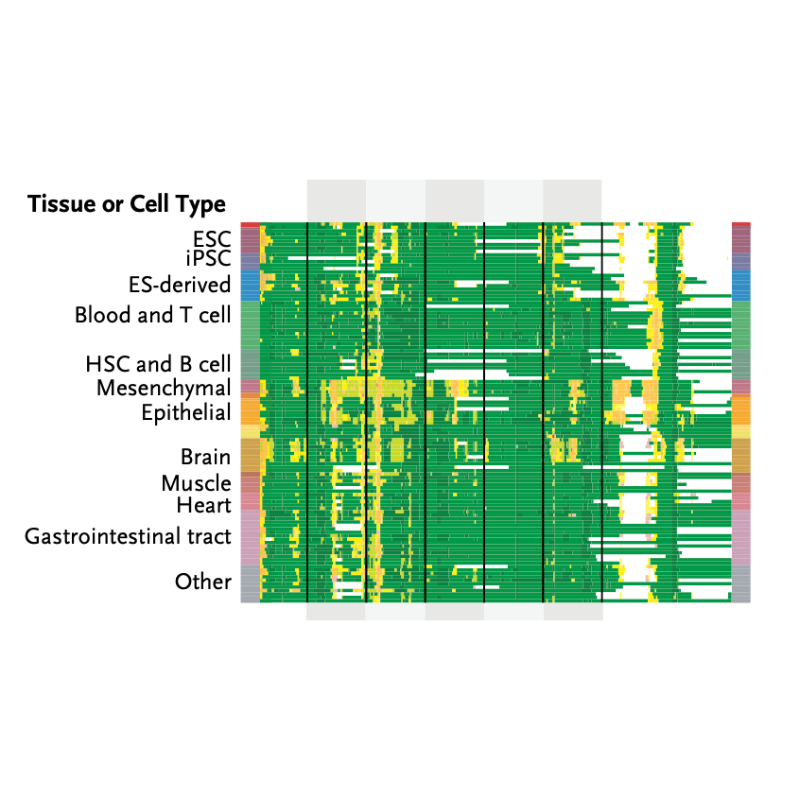 |
149.
FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. (pdf)
Journal: The New England Journal Of Medicine, 2015 Sep 3 doi: 10.1056/NEJMoa1502214 PMID:26287746 PMCID: PMC4959911 |
 |
143.
Alzheimer's loci: epigenetic associations and interaction with genetic factors. (pdf)
Journal: Annals Of Clinical And Translational Neurology, 2015 Jun doi: 10.1002/acn3.201 PMID:26125039 PMCID: PMC4479524 |
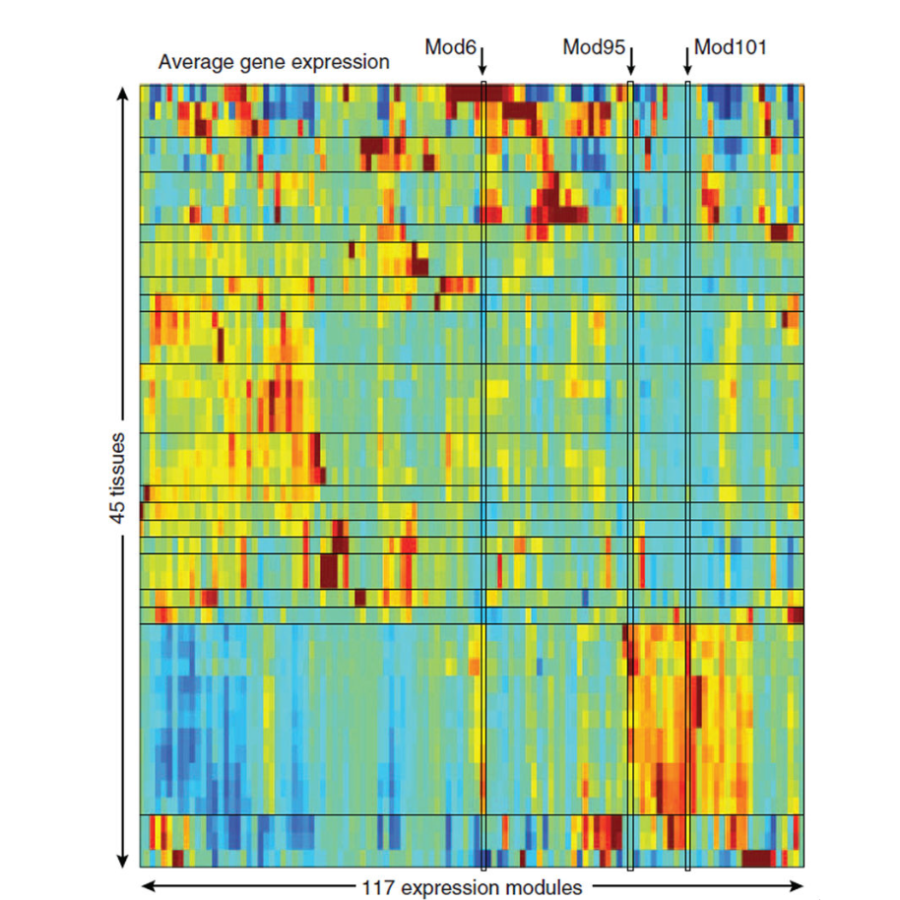 |
141.
Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. (pdf)
Journal: Science, 2015 May 8 doi: 10.1126/science.1262110 PMID:25954001 PMCID: PMC4547484 |
 |
135.
Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. (pdf)
Journal: Nature Genetics, 2015 Apr doi: 10.1038/ng.3245 PMID:25751624 PMCID: PMC4380767 |
 |
133.
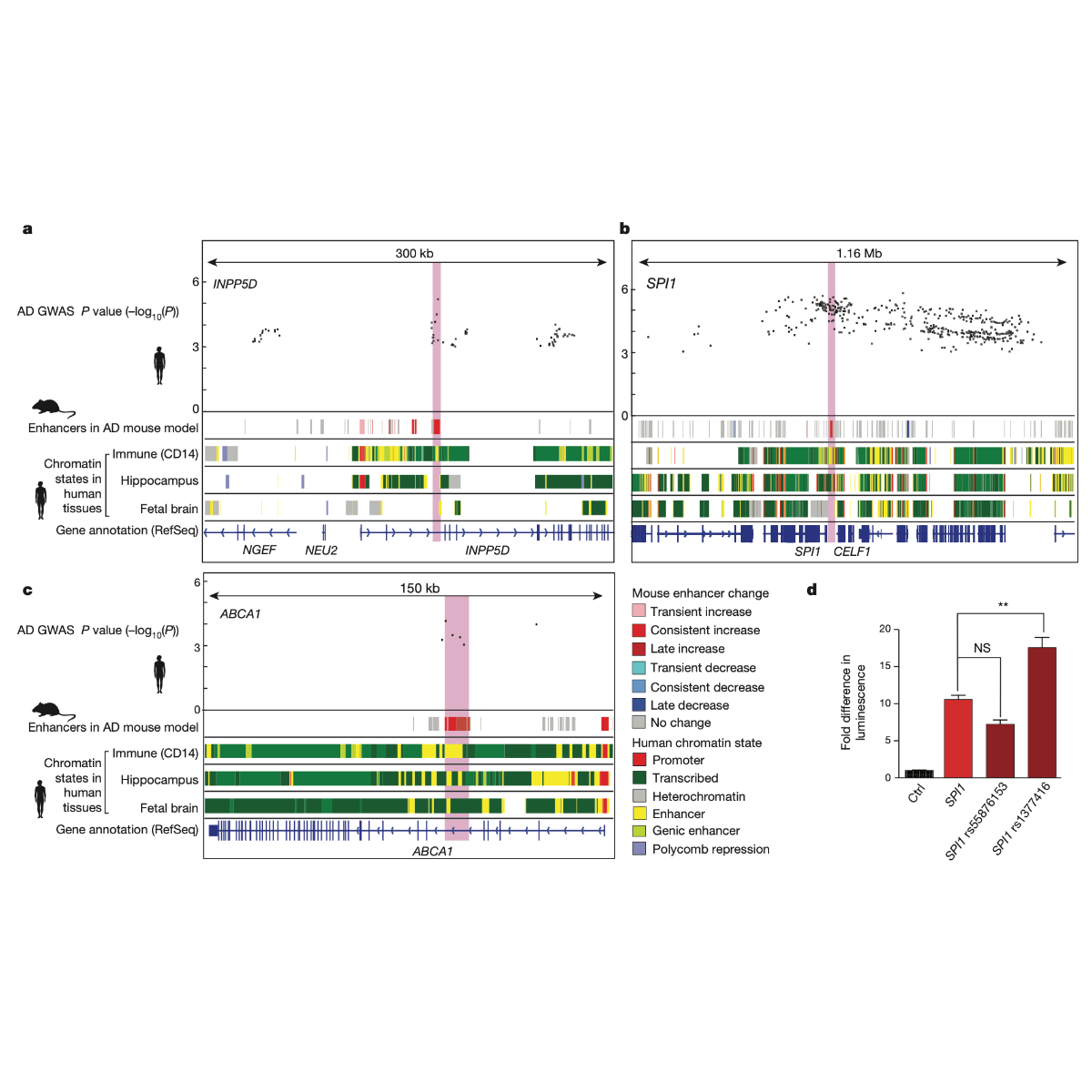
Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. (pdf)
Journal: Nature, 2015 Feb 19 doi: 10.1038/nature14252 PMID:25693568 PMCID: PMC4530583 |
 |
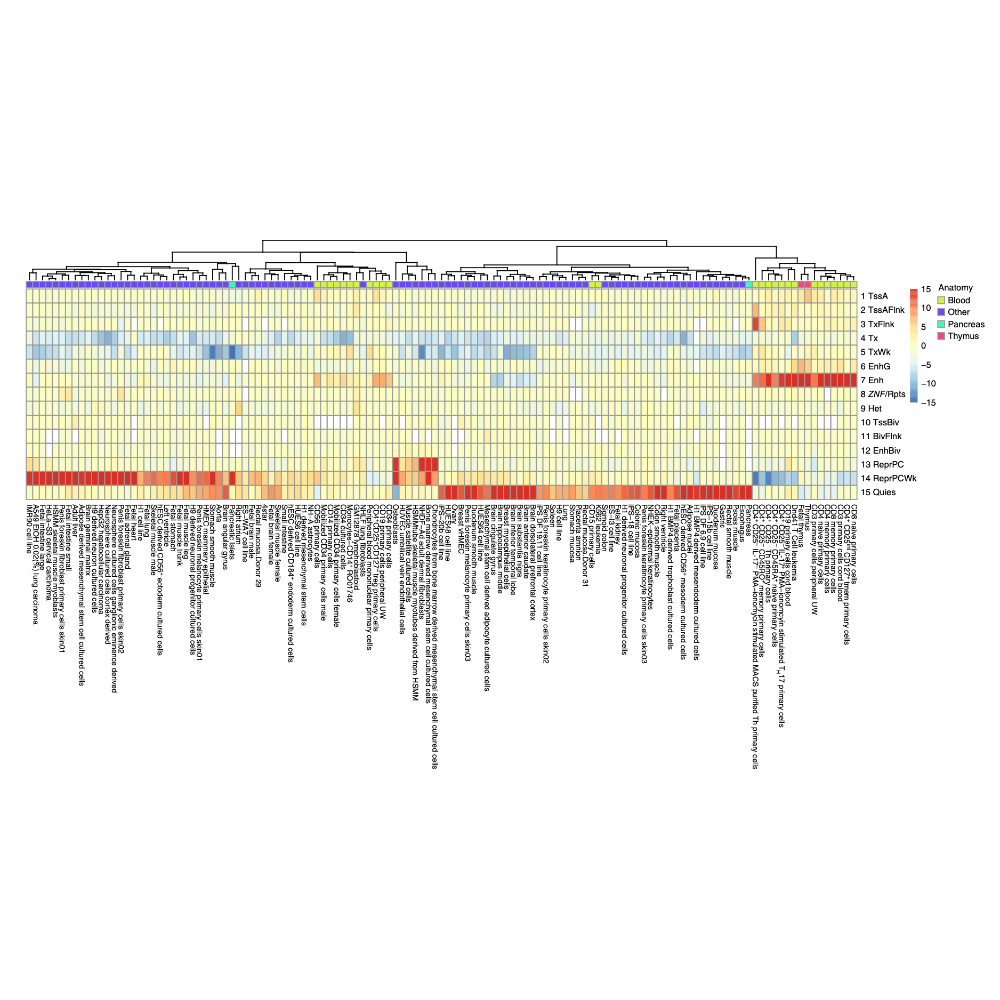
132.
Integrative analysis of 111 reference human epigenomes. (pdf)
Journal: Nature, 2015 Feb 19 doi: 10.1038/nature14248 PMID:25693563 PMCID: PMC4530010 |
 |
125.
CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. (pdf)
Journal: Proceedings Of The National Academy Of Sciences Of The United States Of America, 2014 Oct 21 doi: 10.1073/pnas.1405266111 PMID:25294932 PMCID: PMC4210312 |
 |
123.
Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. (pdf)
Journal: Nature Neuroscience, 2014 Sep doi: 10.1038/nn.3786 PMID:25129075 PMCID: PMC4292795 |
 |
117.
Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. (pdf)
Journal: Nature Genetics, 2014 Aug doi: 10.1038/ng.3014 PMID:24952745 PMCID: PMC4124521 |
 |
110.
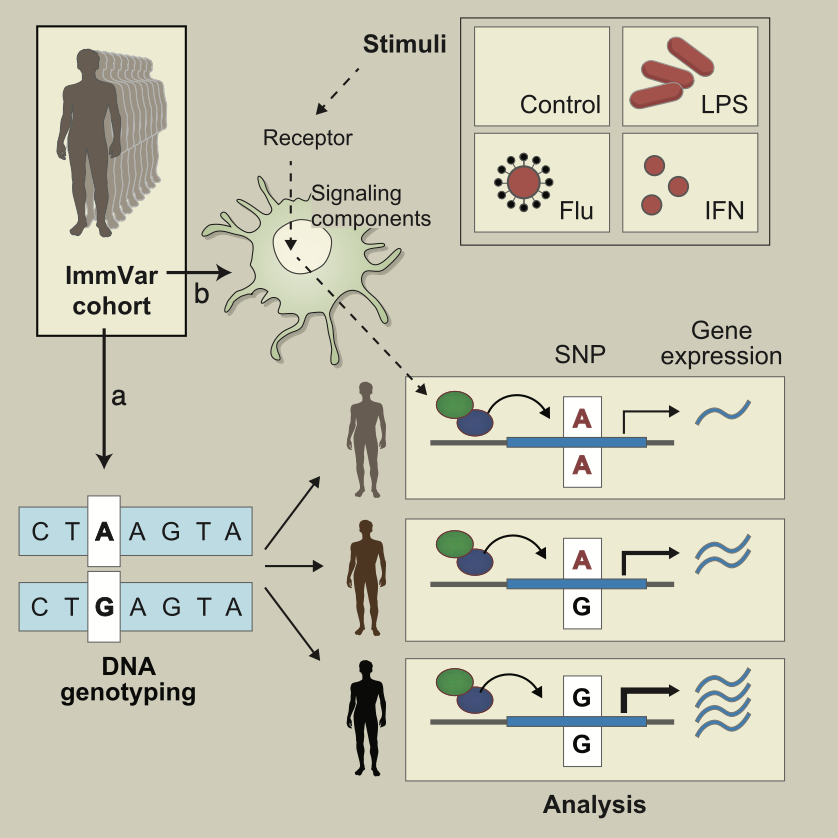
Common genetic variants modulate pathogen-sensing responses in human dendritic cells. (pdf)
Journal: Science, 2014 Mar 7 doi: 10.1126/science.1246980 PMID:24604203 PMCID: PMC4124741 |
 |
101.
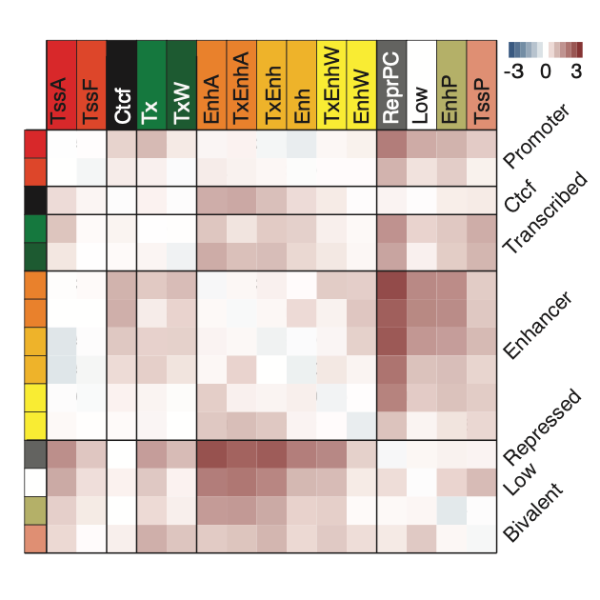
Extensive variation in chromatin states across humans. (pdf)
Journal: Science, 2013 Nov 8 doi: 10.1126/science.1242510 PMID:24136358 PMCID: PMC4075767 |
 |
94.
The Genotype-Tissue Expression (GTEx) project. (pdf)
Journal: Nature Genetics, 2013 Jun doi: 10.1038/ng.2653 PMID:23715323 PMCID: PMC4010069 |
 |
92.
Response to comment on "Evidence of abundant purifying selection in humans for recently acquired regulatory functions". (pdf)
Journal: Science, 2013 May 10 doi: 10.1126/science.1233366 PMID:23661743 PMCID: PMC4131756 |
 |
80.
Interpreting noncoding genetic variation in complex traits and human disease. (pdf)
Journal: Nature Biotechnology, 2012 Nov doi: 10.1038/nbt.2422 PMID:23138309 PMCID: PMC3703467 |
 |
77.
Evidence of abundant purifying selection in humans for recently acquired regulatory functions. (pdf)
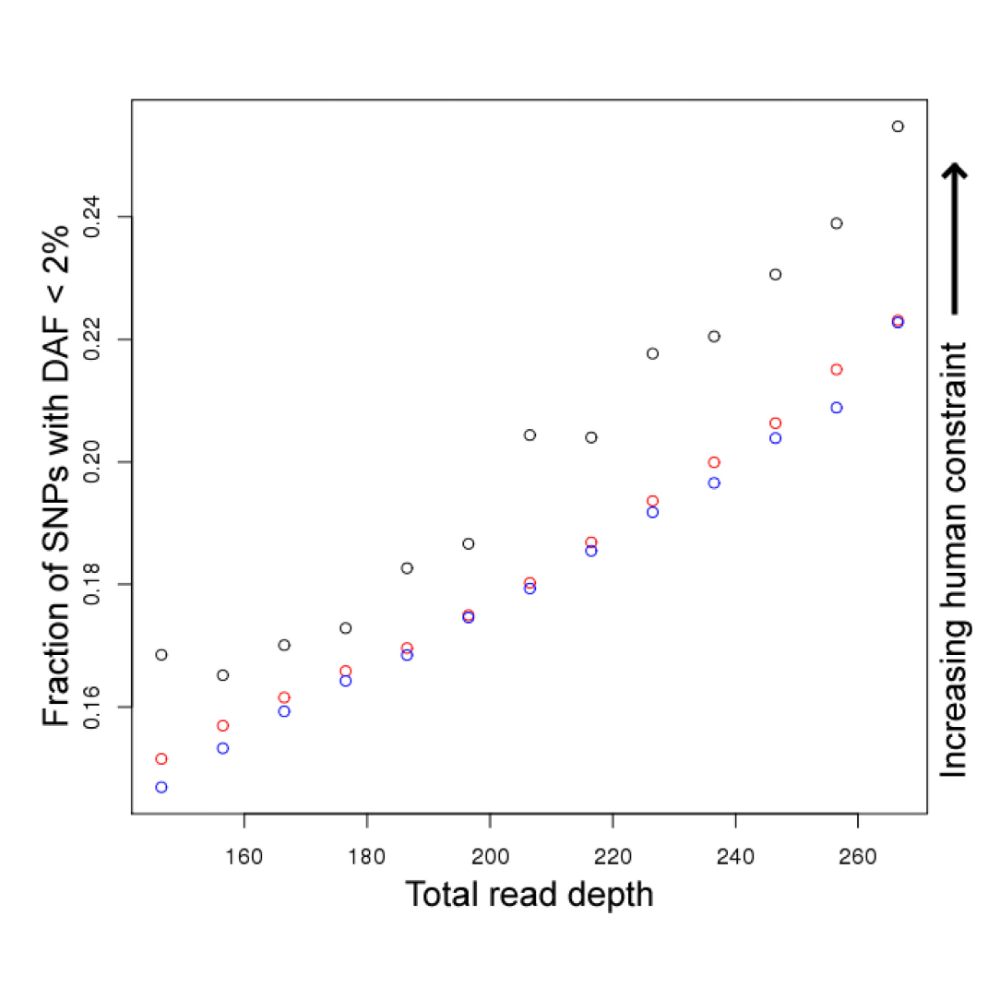
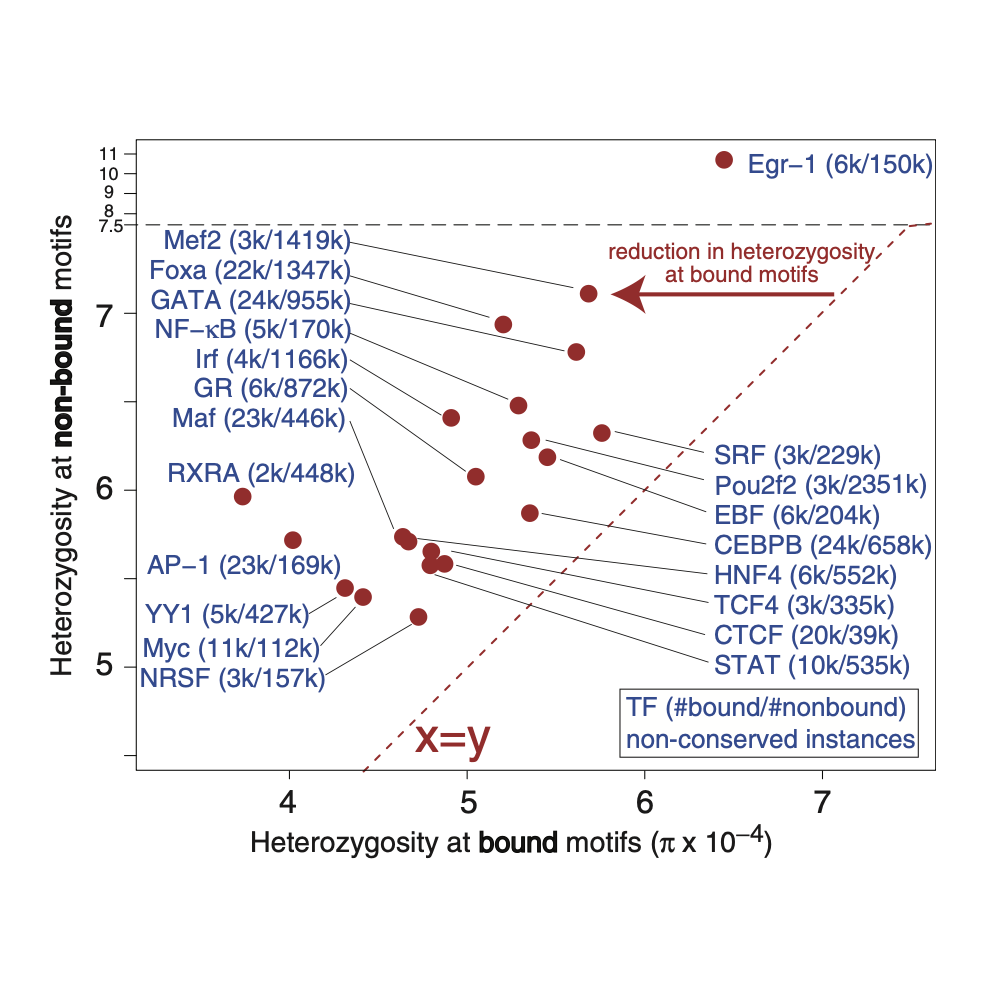
Journal: Science, 2012 Sep 28 doi: 10.1126/science.1225057 PMID:22956687 PMCID: PMC4104271 |
 |
63.
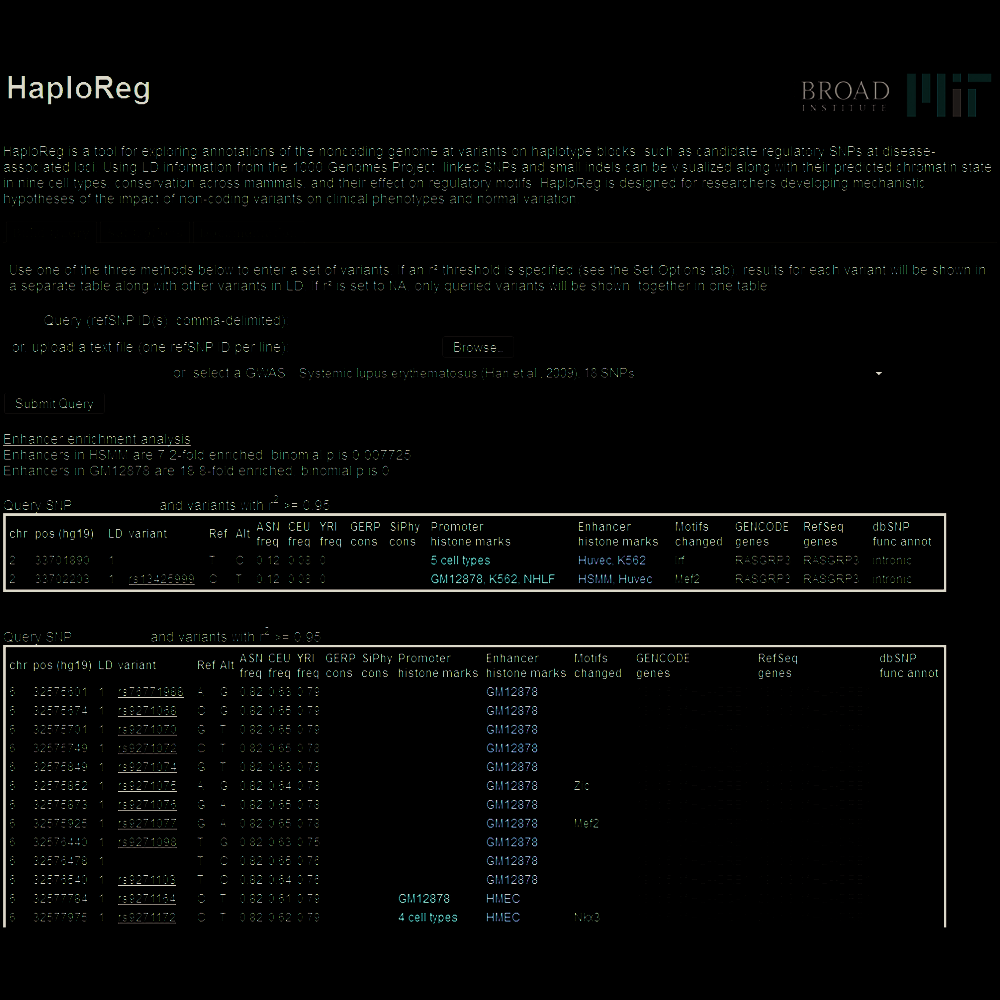
HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. (pdf)
Journal: Nucleic Acids Research, 2012 Jan doi: 10.1093/nar/gkr917 PMID:22064851 PMCID: PMC3245002 |
 |
62.
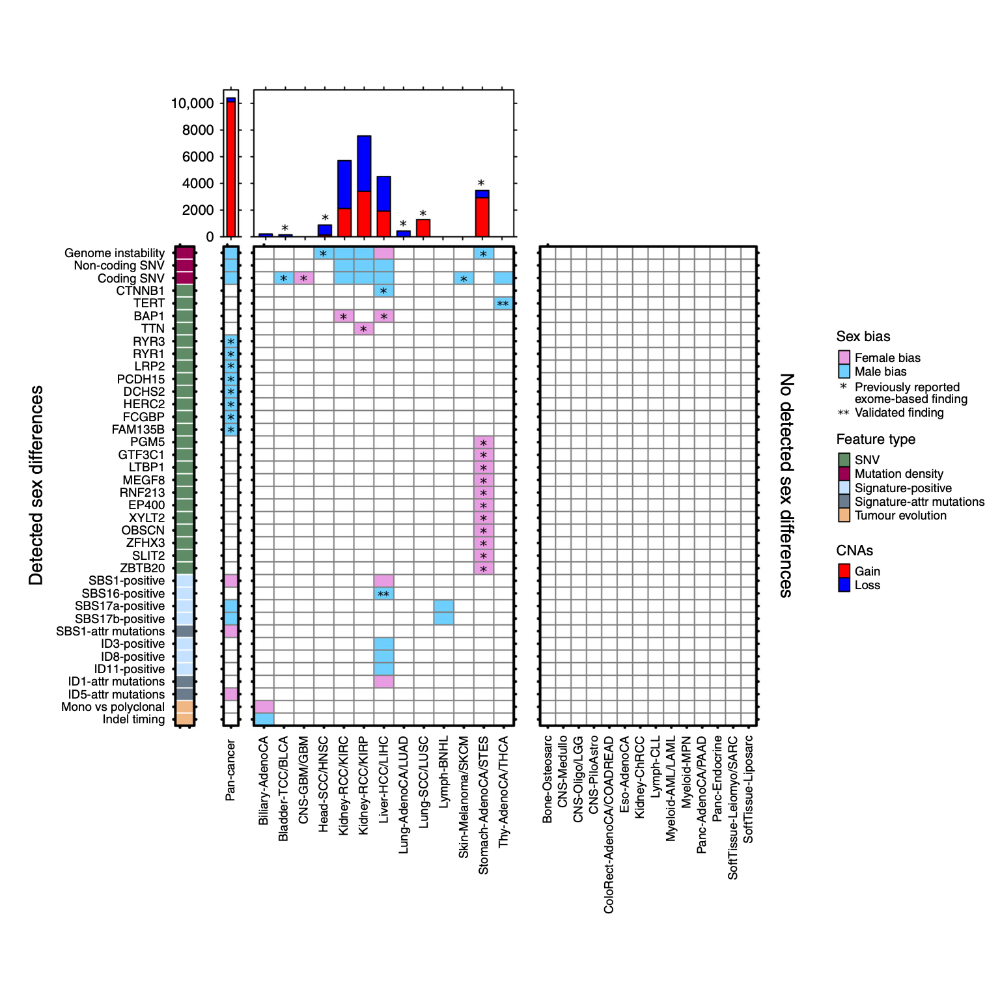
Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. (pdf)
Journal: Plos Genetics, 2012 doi: 10.1371/journal.pgen.1002654 PMID:22570617 PMCID: PMC3343074 |
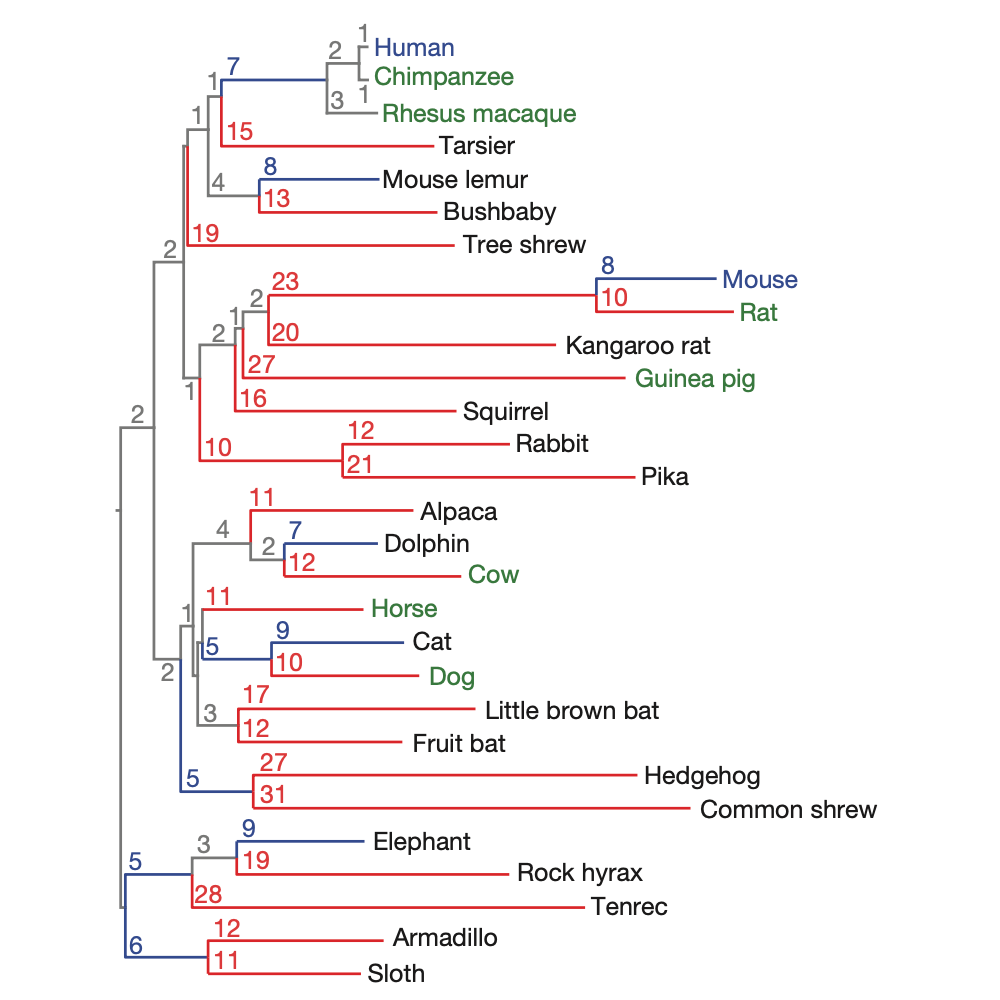 |
57.
A high-resolution map of human evolutionary constraint using 29 mammals. (pdf)
Journal: Nature, 2011 Oct 12 doi: 10.1038/nature10530 PMID:21993624 PMCID: PMC3207357 |
 |
50.
Mapping and analysis of chromatin state dynamics in nine human cell types. (pdf)
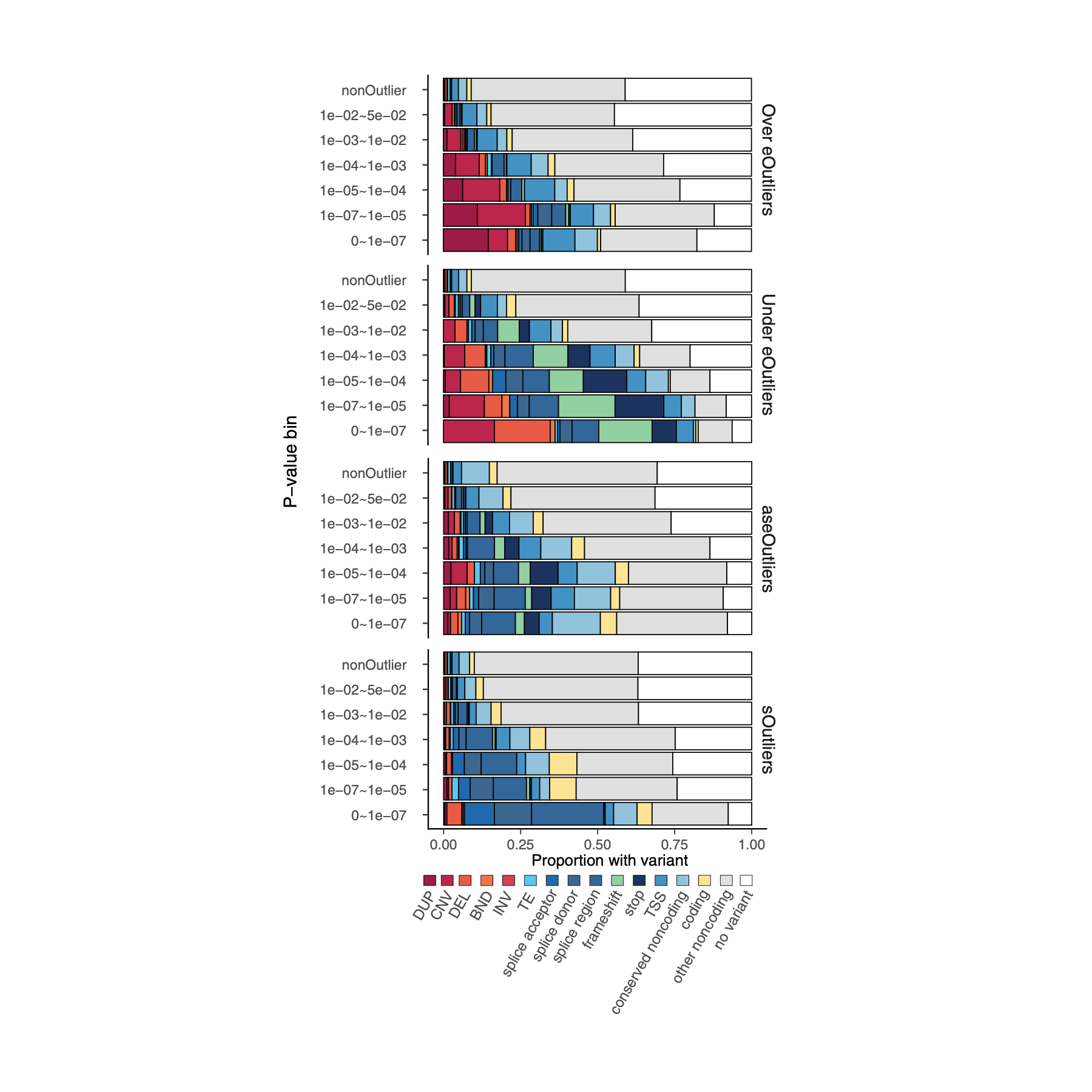
Journal: Nature, 2011 May 5 doi: 10.1038/nature09906 PMID:21441907 PMCID: PMC3088773 |
Genome Interpretation
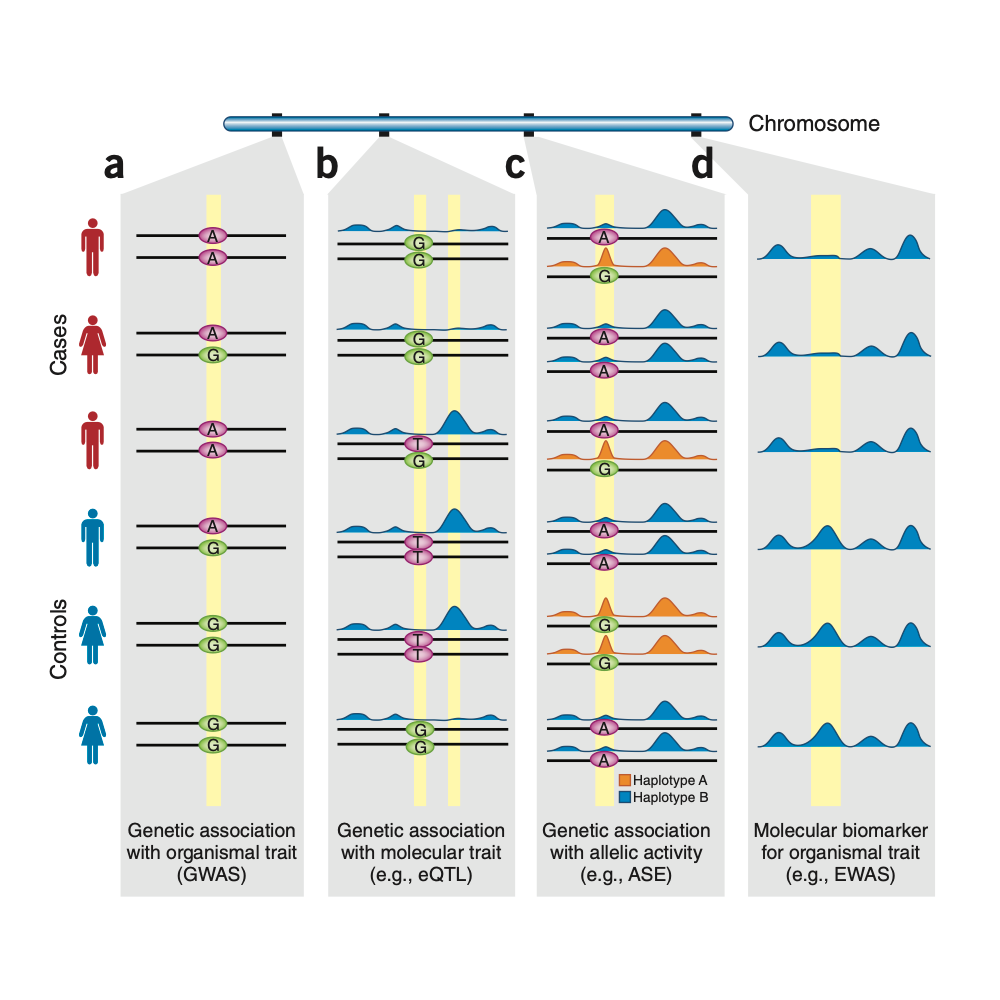
To recognize the molecular basis of human biology and disease, we need a comprehensive understanding of the human genome. This requires computational methods for genome interpretation which can systematically intepret the functional elements encoded in the 4-letter DNA code. For this purpose, we have developed methods for the comprehensive annotation of proteins, RNAs and regulatory control elements encoded in the human genome. While most of these genomic elements lack distinguishing sequence features for their de novo recognition in a single genome, comparison of multiple related species provides a new evolutionary lens for genome interpretation. Exploiting genome-wide comparative genomics datasets, we recognize specific patterns of evolutionary change, or 'evolutionary signatures,' associated with each class of functional elements, and dictated by the specific constraints unique to each type of function. We have used these signatures in the human, fly, and yeast genomes to recognize protein-coding genes and exons, RNA genes and structures, microRNAs and their targets, and several types of regulatory elements, revealing thousands of new elements, and many novel biological insights.
 |
269.
A wealth of discovery built on the Human Genome Project - by the numbers. (pdf)
Journal: Nature, 2021 Feb doi: 10.1038/d41586-021-00314-6 PMID:33568828 |
 |
268.
Regulatory genomic circuitry of human disease loci by integrative epigenomics. (pdf)
Journal: Nature, 2021 Feb doi: 10.1038/s41586-020-03145-z PMID:33536621 PMCID: PMC7875769 |
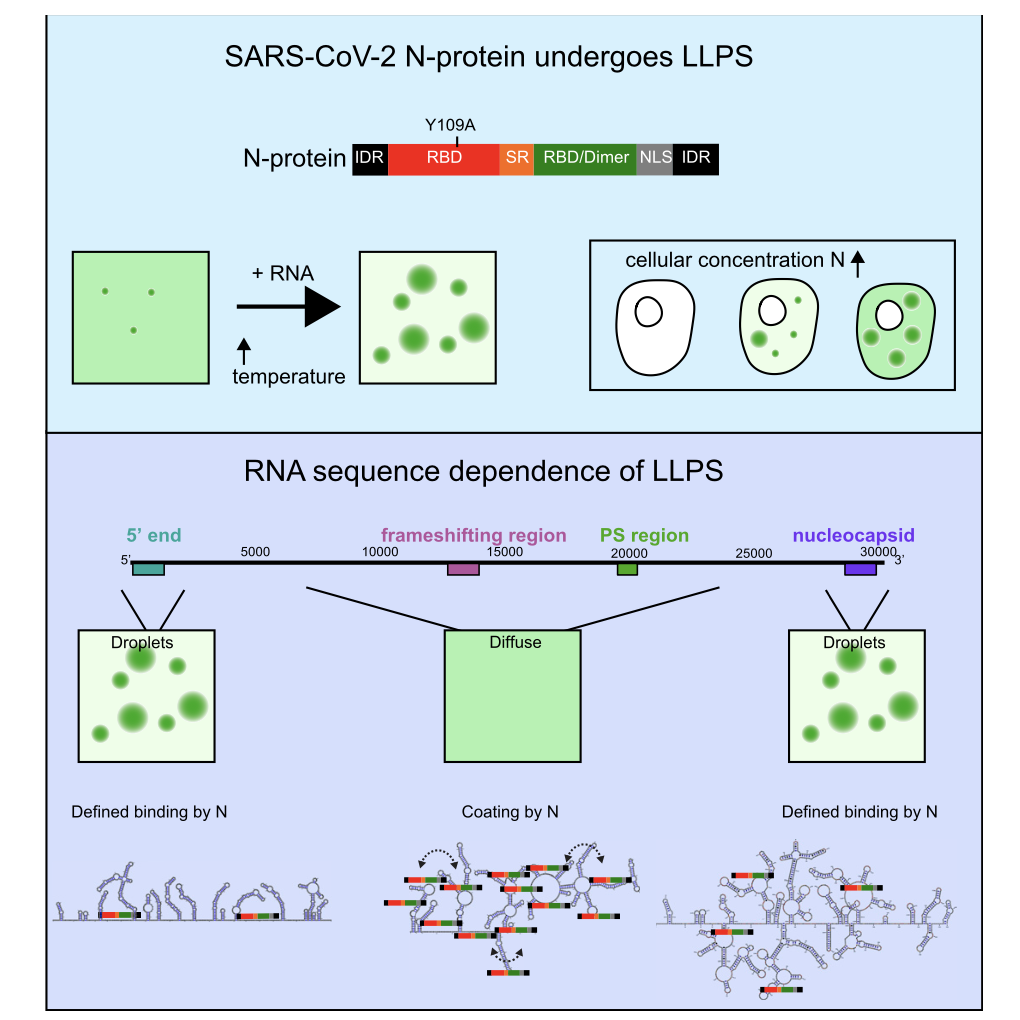 |
265.
Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. (pdf)
Journal: Molecular Cell, 2020 Dec 17 doi: 10.1016/j.molcel.2020.11.041 PMID:33290746 PMCID: PMC7691212 |
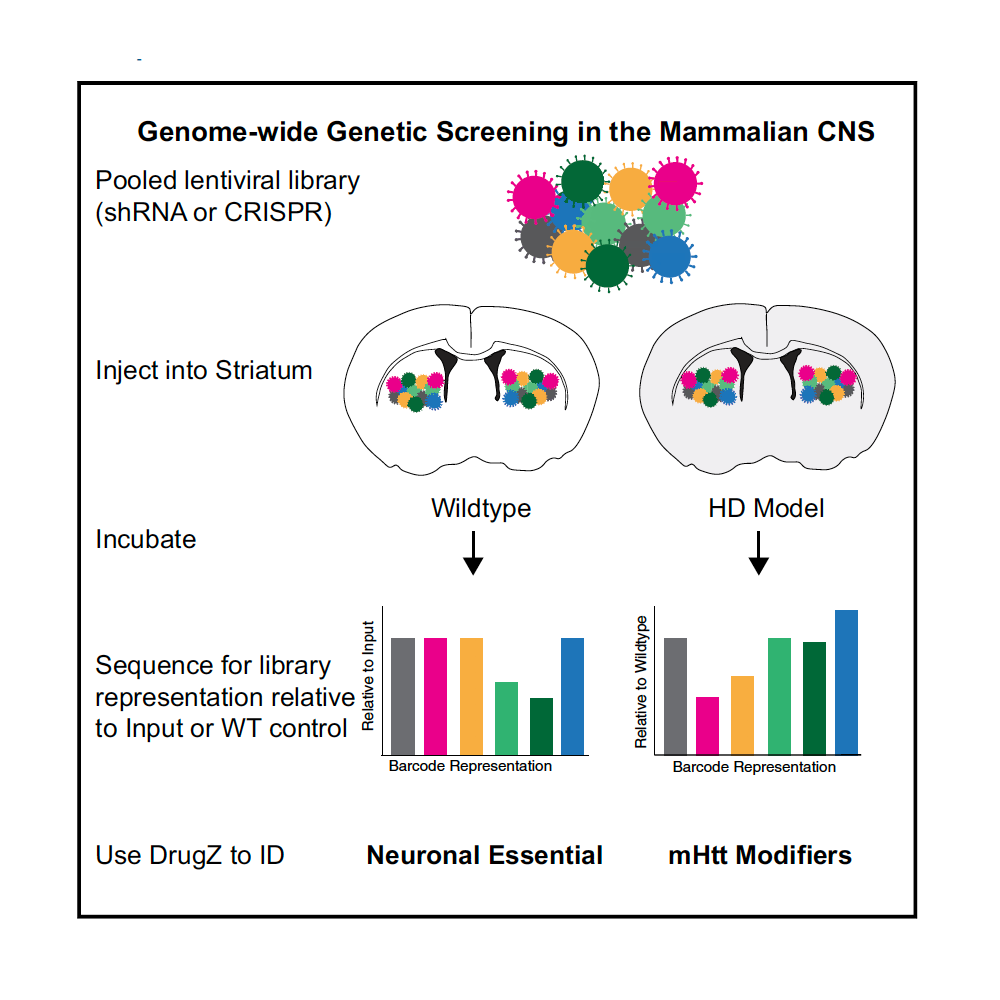 |
238.
Genome-wide In Vivo CNS Screening Identifies Genes that Modify CNS Neuronal Survival and mHTT Toxicity. (pdf)
Journal: Neuron, 2020 Apr 8 doi: 10.1016/j.neuron.2020.01.004 PMID:32004439 PMCID: PMC7181458 |
 |
233.
Comprehensive molecular characterization of mitochondrial genomes in human cancers. (pdf)
Journal: Nature Genetics, 2020 Mar doi: 10.1038/s41588-019-0557-x PMID:32024997 PMCID: PMC7058535 |
 |
232.
Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. (pdf)
Journal: Nature Genetics, 2020 Mar doi: 10.1038/s41588-019-0576-7 PMID:32025003 PMCID: PMC7058534 |
 |
231.
Butler enables rapid cloud-based analysis of thousands of human genomes. (pdf)
Journal: Nature Biotechnology, 2020 Mar doi: 10.1038/s41587-019-0360-3 PMID:32024987 PMCID: PMC7062635 |
 |
230.
Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. (pdf)
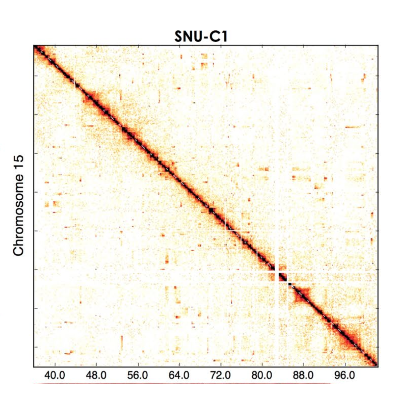
Journal: Nature Genetics, 2020 Mar doi: 10.1038/s41588-019-0564-y PMID:32024999 PMCID: PMC7058537 |
 |
229.
Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. (pdf)
Journal: Nature Genetics, 2020 Mar doi: 10.1038/s41588-019-0562-0 PMID:32024998 PMCID: PMC7058536 |
 |
228.
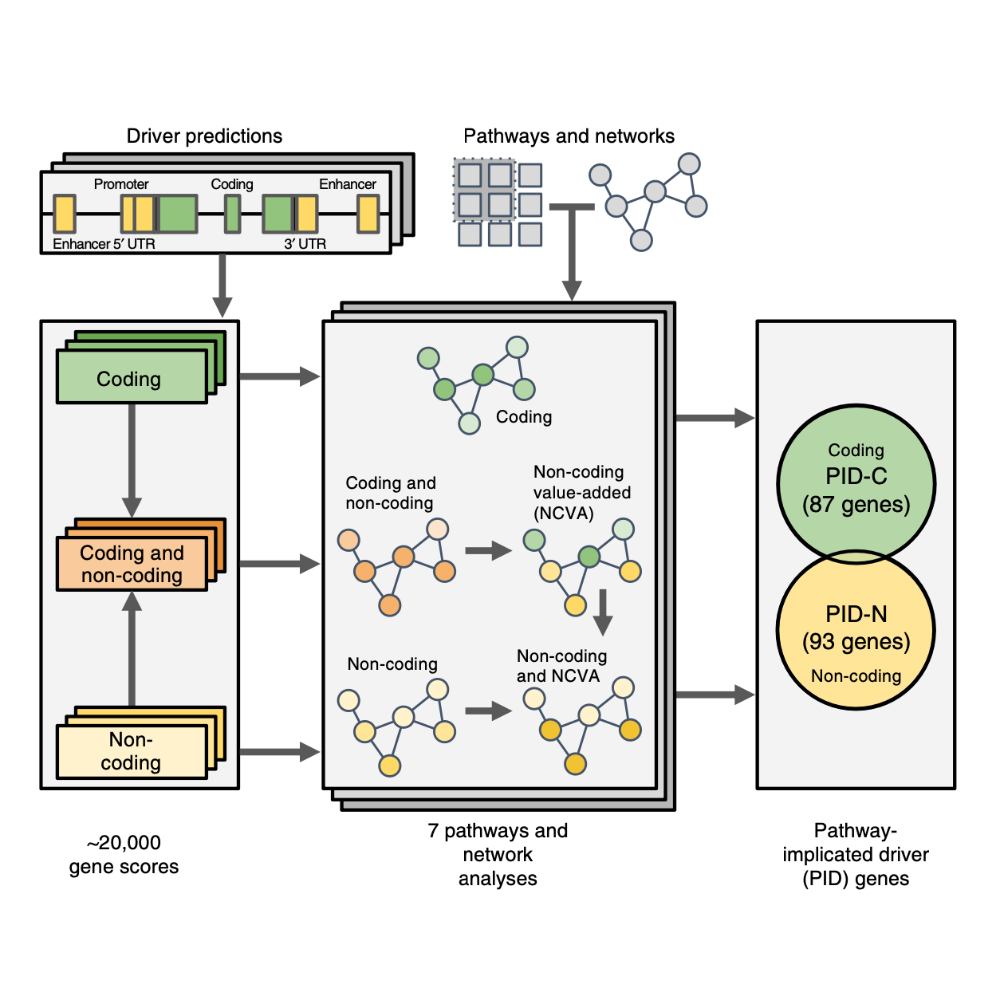
Pathway and network analysis of more than 2500 whole cancer genomes. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-020-14367-0 PMID:32024854 PMCID: PMC7002574 |
 |
224.
High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-019-13885-w PMID:32024823 PMCID: PMC7002524 |
 |
220.
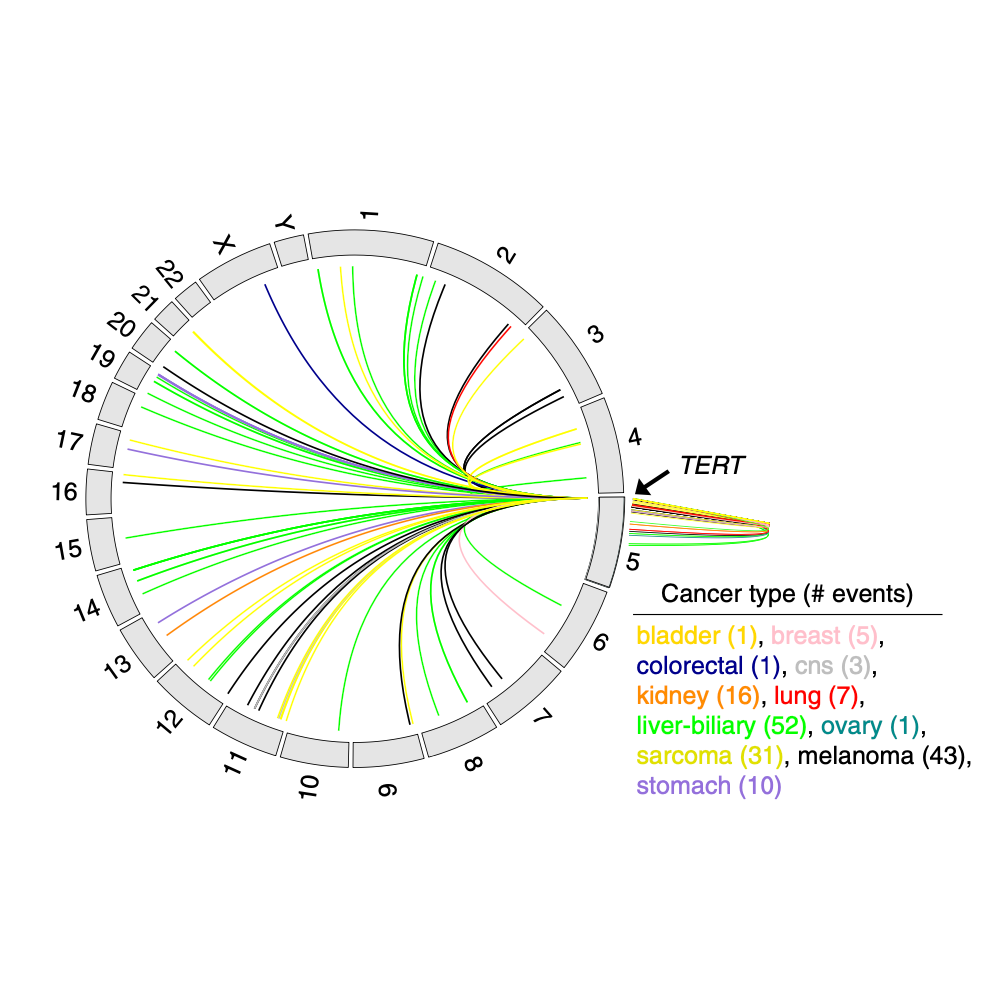
Genomic footprints of activated telomere maintenance mechanisms in cancer. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-019-13824-9 PMID:32024817 PMCID: PMC7002710 |
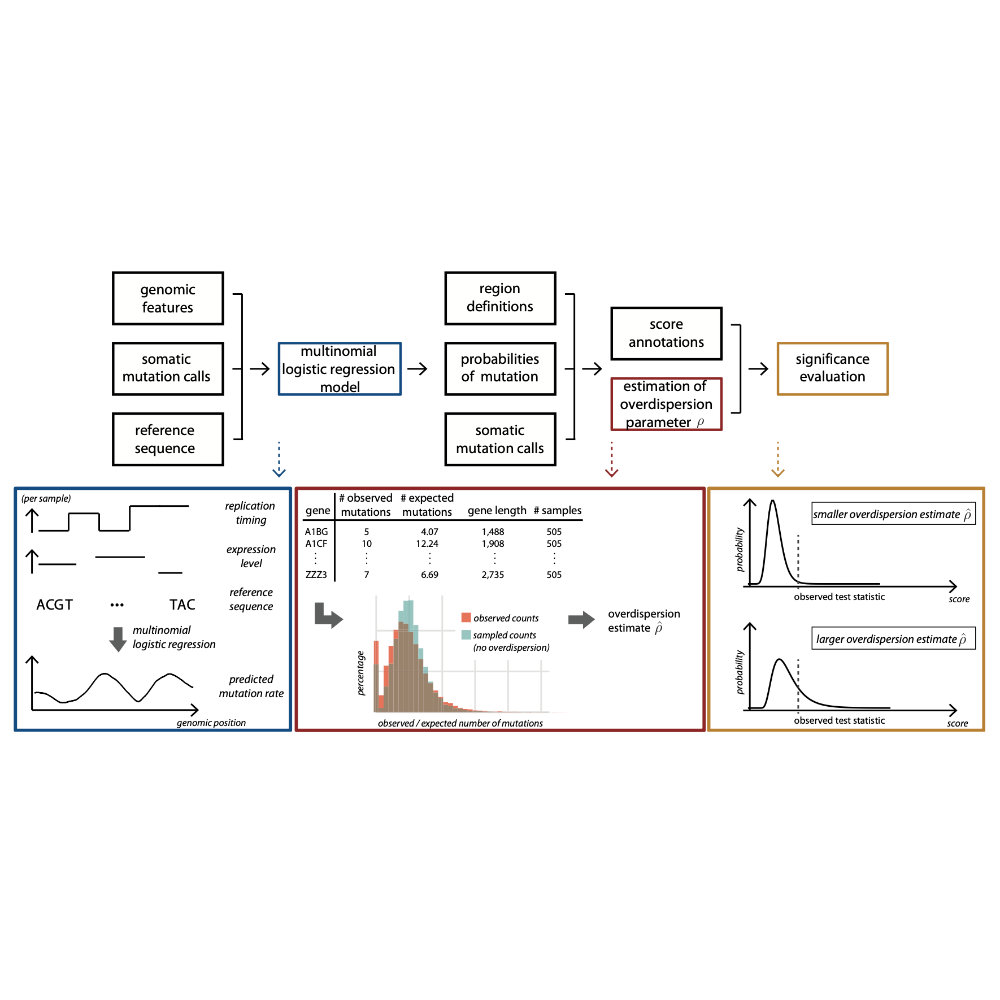 |
197.
ncdDetect2: improved models of the site-specific mutation rate in cancer and driver detection with robust significance evaluation. (pdf)
Journal: Bioinformatics, 2019 Jan 15 doi: 10.1093/bioinformatics/bty511 PMID:29945188 PMCID: PMC6330011 |
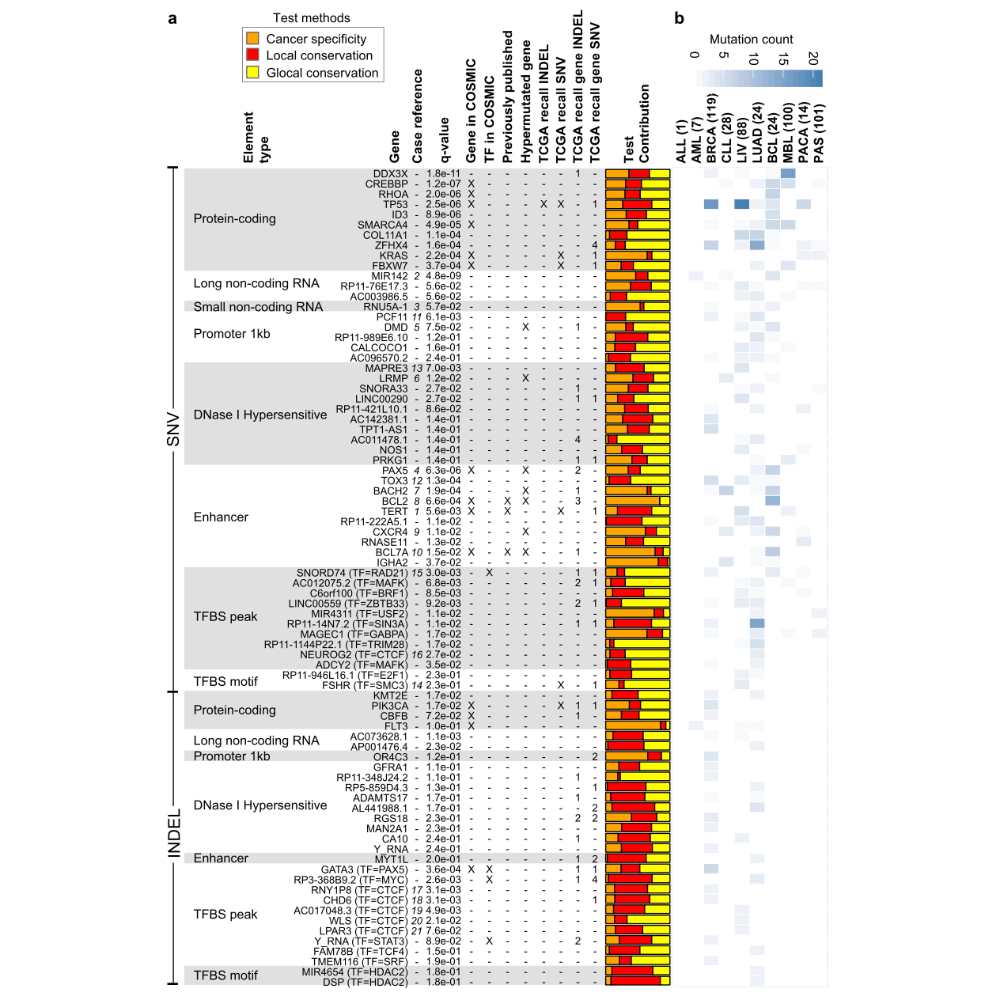 |
179.
Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. (pdf)
Journal: Npj Genomic Medicine, 2018 doi: 10.1038/s41525-017-0040-5 PMID:29354286 PMCID: PMC5765157 |
 |
132.
Integrative analysis of 111 reference human epigenomes. (pdf)
Journal: Nature, 2015 Feb 19 doi: 10.1038/nature14248 PMID:25693563 PMCID: PMC4530010 |
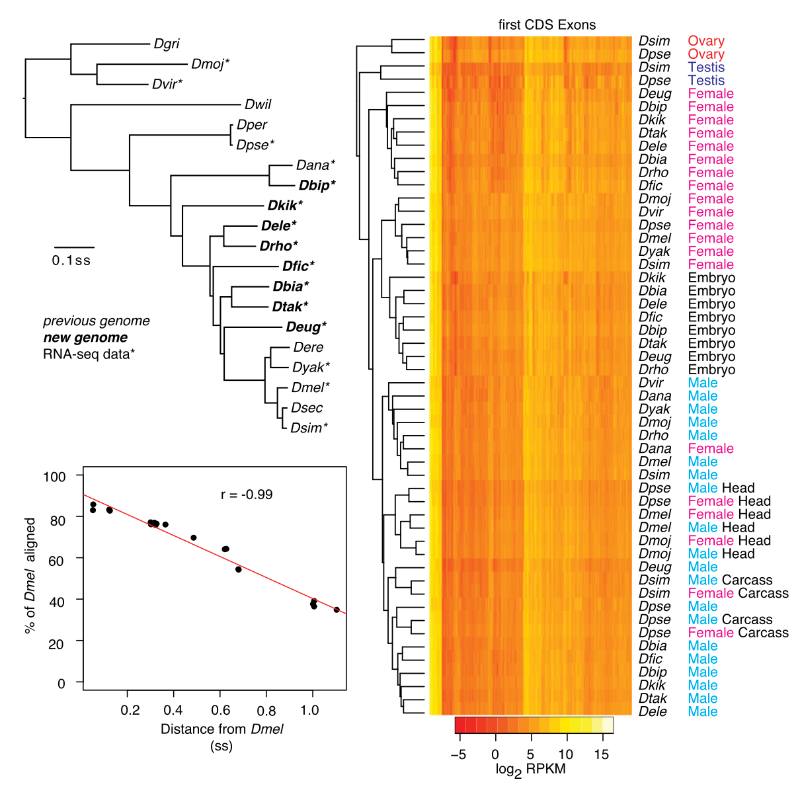 |
115.
Comparative validation of the D. melanogaster modENCODE transcriptome annotation. (pdf)
Journal: Genome Research, 2014 Jul doi: 10.1101/gr.159384.113 PMID:24985915 PMCID: PMC4079975 |
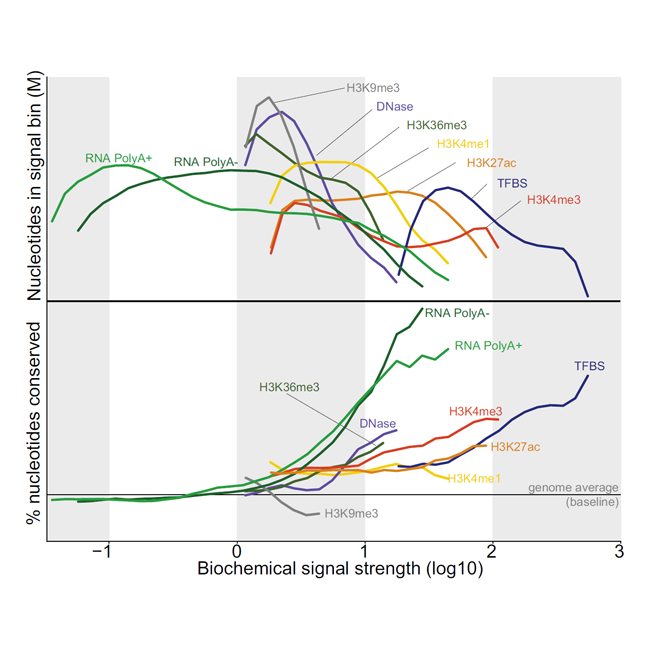 |
112.
Defining functional DNA elements in the human genome. (pdf)
Journal: Proceedings Of The National Academy Of Sciences Of The United States Of America, 2014 Apr 29 doi: 10.1073/pnas.1318948111 PMID:24753594 PMCID: PMC4035993 |
 |
75.
An integrated encyclopedia of DNA elements in the human genome. (pdf)
Journal: Nature, 2012 Sep 6 doi: 10.1038/nature11247 PMID:22955616 PMCID: PMC3439153 |
 |
49.
A user's guide to the encyclopedia of DNA elements (ENCODE). (pdf)
Journal: Plos Biology, 2011 Apr doi: 10.1371/journal.pbio.1001046 PMID:21526222 PMCID: PMC3079585 |
 |
42.
Identification of functional elements and regulatory circuits by Drosophila modENCODE. (pdf)
Journal: Science, 2010 Dec 24 doi: 10.1126/science.1198374 PMID:21177974 PMCID: PMC3192495 |
 |
33.
Unlocking the secrets of the genome. (pdf)
Journal: Nature, 2009 Jun 18 doi: 10.1038/459927a PMID:19536255 PMCID: PMC2843545 |
 |
15.
Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. (pdf)
Journal: Nature, 2007 Nov 8 doi: 10.1038/nature06340 PMID:17994088 PMCID: PMC2474711 |
 |
11.
Genome sequence, comparative analysis and haplotype structure of the domestic dog. (pdf)
Journal: Nature, 2005 Dec 8 doi: 10.1038/nature04338 PMID:16341006 |
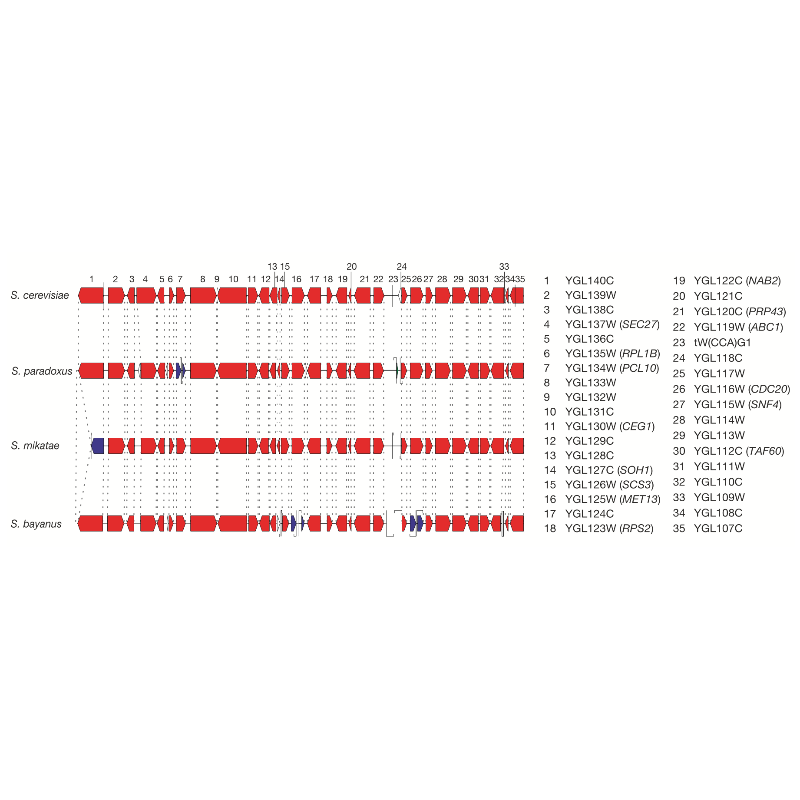 |
2.
Sequencing and comparison of yeast species to identify genes and regulatory elements. (pdf)
Journal: Nature, 2003 May 15 doi: 10.1038/nature01644 PMID:12748633 |
Brain Genomics
A major focus of our lab has been the single-cell dissection of brain disorders, focusing on Alzheimer's Disease, Schizophrenia, ALS, FTD, Hungtington's Disease, PTSD, and diverse psychiatric and neurodegenerative disorders. We have shown that genetic variants associated with Alzheimer's disease localize specifically within immune enhancers, indicating that neuroinflammation and microglial processes might be the first line of defense against Alzheimer's and providing a new therapeutic hypothesis that may precede Amyoid and Tangles. We have provided the first single-cell dissection of Schizophrenia, revealing a complex interplay of multiple cell types, and revealing a common synaptic/neurodevelopmental axis of common transcription factors. We generated the first single-cell dissection of ALS, FTD, Bipolar, and Huntington's Disease, providing numerous insights on their common and distinct underpinnings. We were able to distinguish multiple subtypes of microglia, revealing their distinct roles in Schizophrenia vs. Alzheimer's disease. We generated the largest resource of single-cell epigenomics in the human brain, helping annotate the distinct genomic circuitry of each major cell type. We're also developing methods for probing the genomic circuitry of brain cells through high-throughput perturbations. This continues to be a strong area of focus of our lab, through multiple fruitful collaborations.
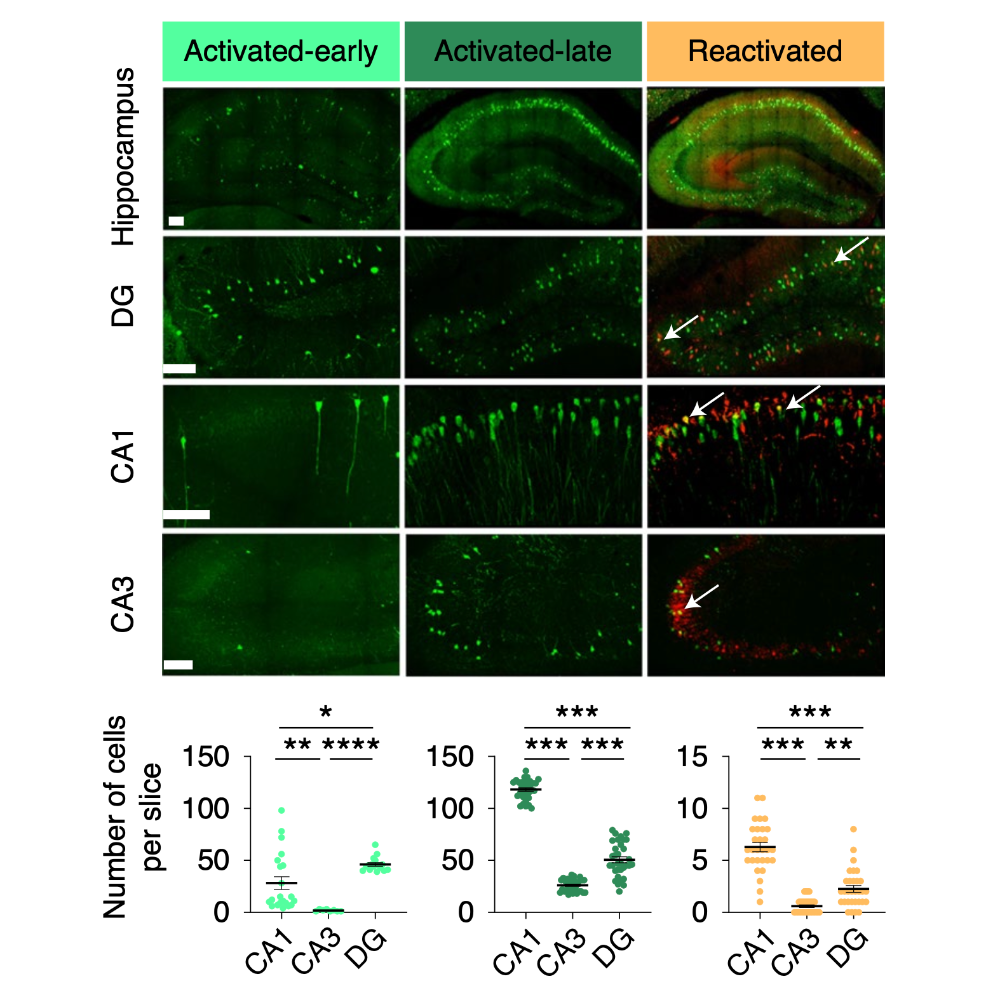 |
263.
Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. (pdf)
Journal: Nature Neuroscience, 2020 Dec doi: 10.1038/s41593-020-00717-0 PMID:33020654 PMCID: PMC7686266 |
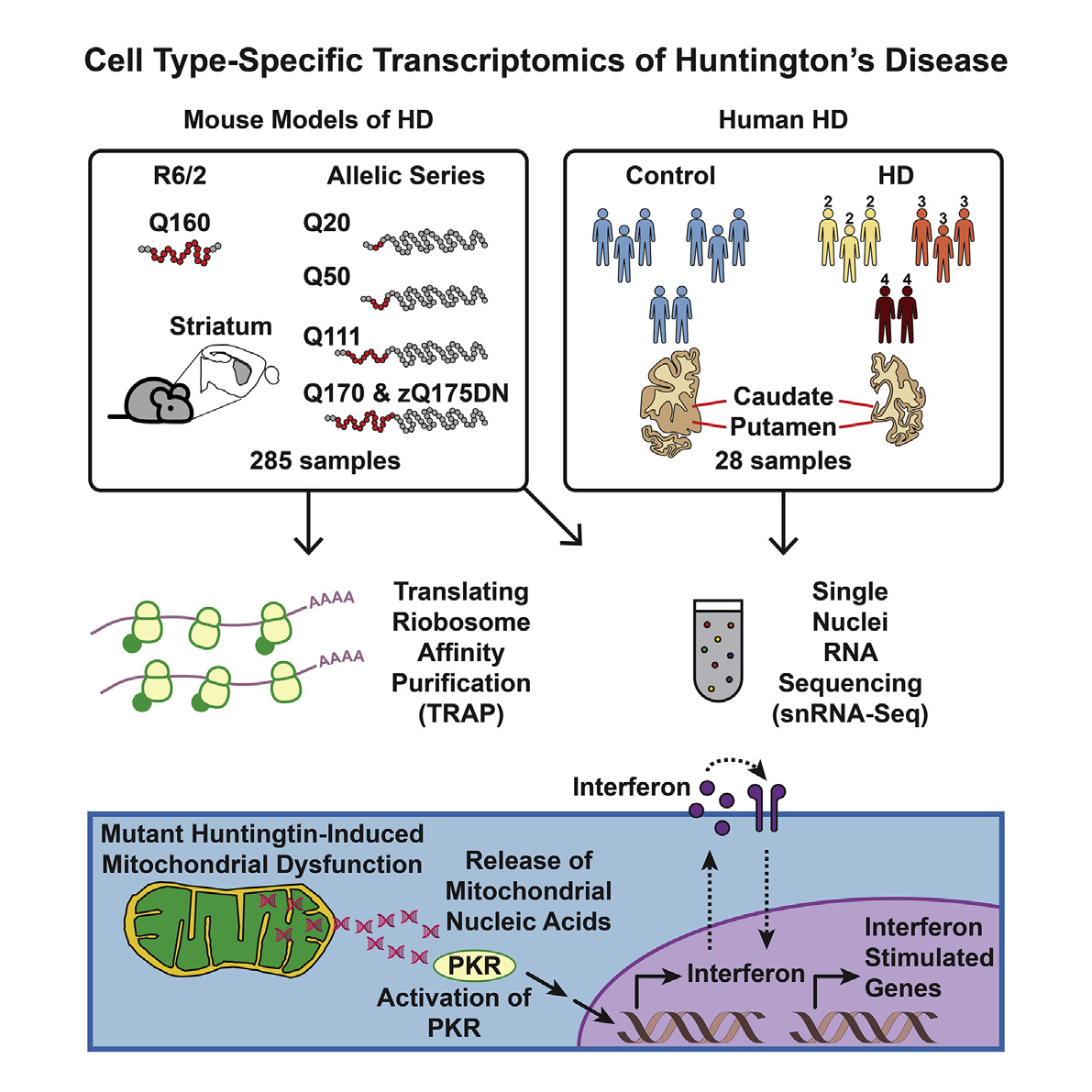 |
249.
Cell Type-Specific Transcriptomics Reveals that Mutant Huntingtin Leads to Mitochondrial RNA Release and Neuronal Innate Immune Activation. (pdf)
Journal: Neuron, 2020 Sep 9 doi: 10.1016/j.neuron.2020.06.021 PMID:32681824 PMCID: PMC7486278 |
 |
248.
Alternatives to amyloid for Alzheimer's disease therapies-a symposium report. (pdf)
Journal: Annals Of The New York Academy Of Sciences, 2020 Sep doi: 10.1111/nyas.14371 PMID:32472577 PMCID: PMC9621603 |
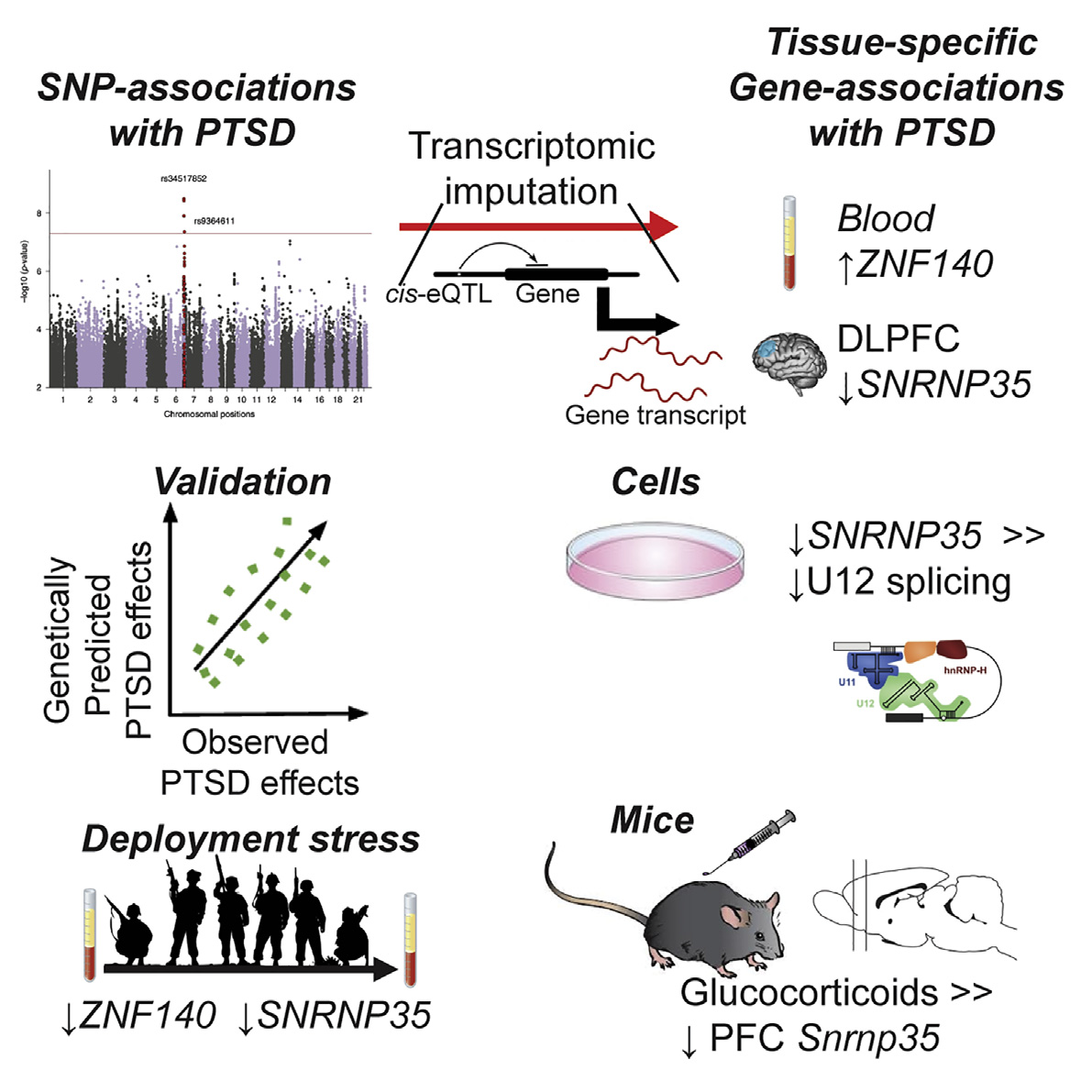 |
242.
Analysis of Genetically Regulated Gene Expression Identifies a Prefrontal PTSD Gene, SNRNP35, Specific to Military Cohorts. (pdf)
Journal: Cell Reports, 2020 Jun 2 doi: 10.1016/j.celrep.2020.107716 PMID:32492425 PMCID: PMC7359754 |
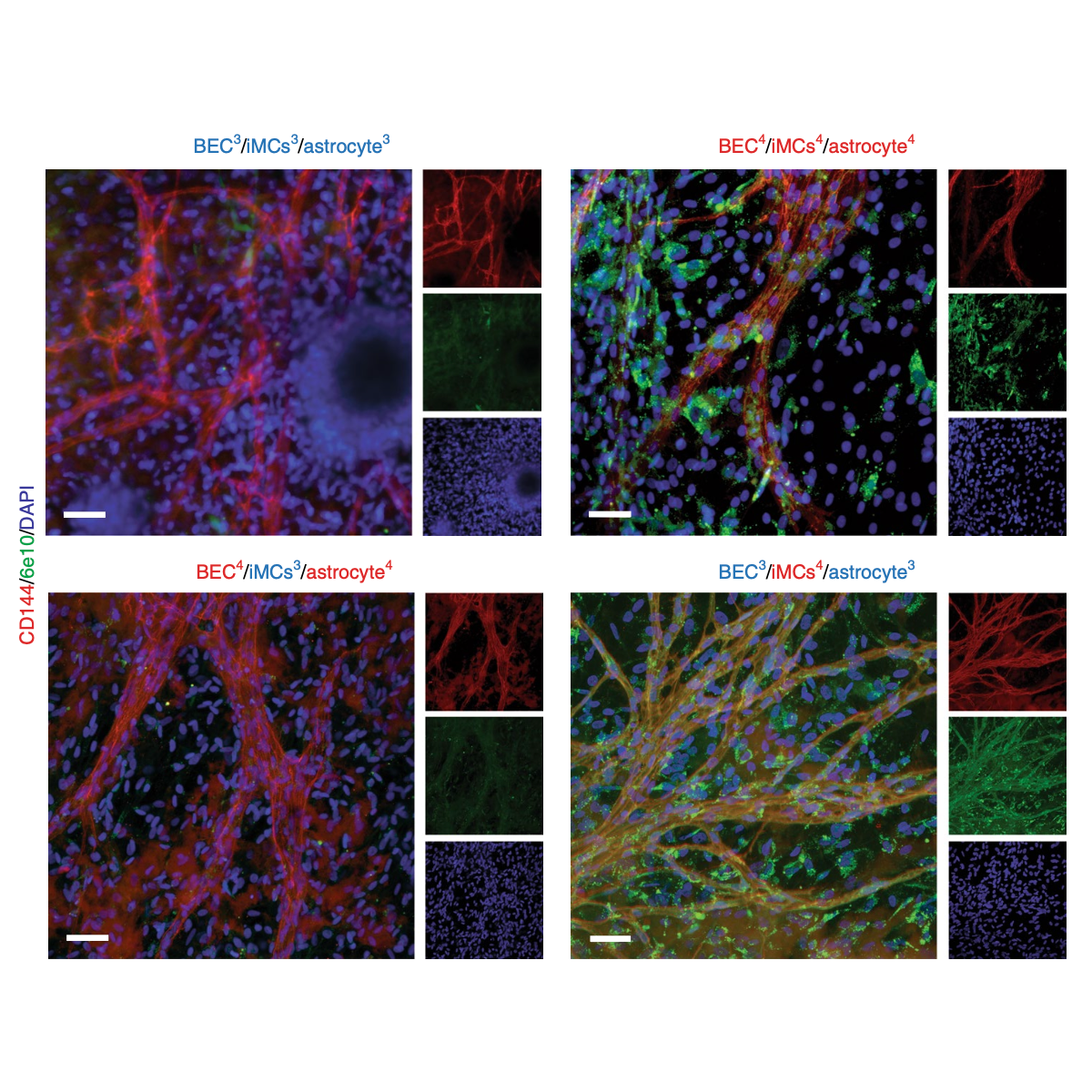 |
241.
Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. (pdf)
Journal: Nature Medicine, 2020 Jun doi: 10.1038/s41591-020-0886-4 PMID:32514169 PMCID: PMC7704032 |
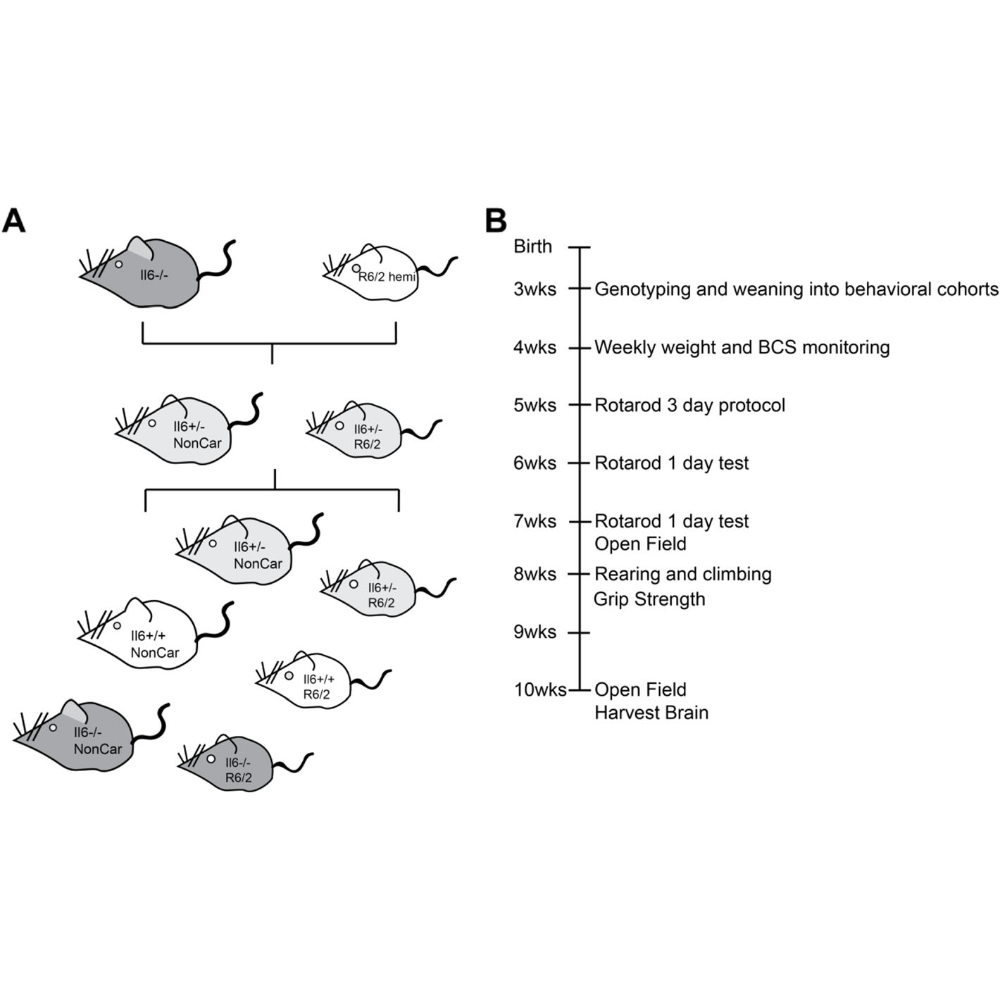 |
240.
Interleukin-6 deficiency exacerbates Huntington's disease model phenotypes. (pdf)
Journal: Molecular Neurodegeneration, 2020 May 24 doi: 10.1186/s13024-020-00379-3 PMID:32448329 PMCID: PMC7247164 |
 |
238.
Genome-wide In Vivo CNS Screening Identifies Genes that Modify CNS Neuronal Survival and mHTT Toxicity. (pdf)
Journal: Neuron, 2020 Apr 8 doi: 10.1016/j.neuron.2020.01.004 PMID:32004439 PMCID: PMC7181458 |
 |
204.
Rate of brain aging and APOE epsilon4 are synergistic risk factors for Alzheimer's disease. (pdf)
Journal: Life Science Alliance, 2019 Jun doi: 10.26508/lsa.201900303 PMID:31133613 PMCID: PMC6537750 |
 |
203.
Single-cell transcriptomic analysis of Alzheimer's disease. (pdf)
Journal: Nature, 2019 Jun doi: 10.1038/s41586-019-1195-2 PMID:31042697 PMCID: PMC6865822 |
 |
143.
Alzheimer's loci: epigenetic associations and interaction with genetic factors. (pdf)
Journal: Annals Of Clinical And Translational Neurology, 2015 Jun doi: 10.1002/acn3.201 PMID:26125039 PMCID: PMC4479524 |
 |
133.
Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. (pdf)
Journal: Nature, 2015 Feb 19 doi: 10.1038/nature14252 PMID:25693568 PMCID: PMC4530583 |
Cancer Genomics
Another major interst of our lab is understanding the genomics, epigenomics, and transcriptomics of cancer progression. Cancer poses many unique challenges by relative to common disease, by tapping into the rare and somatic end of the allele frequency spectrum (rather than common alleles in GWAS), by exploiting positive selection for change (rather than purifying selection observed against most disease variants), by reprogramming cellular identity (rather than relying on normal tissue-specific regulatory programs), the challenge of detecting its epigenomic and transcriptional state in inaccessible organs (rather than having access to post-mortem samples for analysis), and by its unique interactions and co-option of immune processes. Our work in cancer brings together the diverse areas of expertise of our lab, across comparative genomics, phylogenomics, and evolution; regulatory genomics and epigenomics; immune circuitry and interactions; and interpretation of coding and non-coding mutations.
 |
270.
Distinct metabolic programs established in the thymus control effector functions of gammadelta T cell subsets in tumor microenvironments. (pdf)
Journal: Nature Immunology, 2021 Feb doi: 10.1038/s41590-020-00848-3 PMID:33462452 PMCID: PMC7610600 |
 |
262.
Plasma-derived extracellular vesicle analysis and deconvolution enable prediction and tracking of melanoma checkpoint blockade outcome. (pdf)
Journal: Science Advances, 2020 Nov doi: 10.1126/sciadv.abb3461 PMID:33188016 PMCID: PMC7673759 |
 |
257.
Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. (pdf)
Journal: Nature Communications, 2020 Sep 21 doi: 10.1038/s41467-020-18151-y PMID:32958763 PMCID: PMC7505971 |
 |
247.
Sex differences in oncogenic mutational processes. (pdf)
Journal: Nature Communications, 2020 Aug 28 doi: 10.1038/s41467-020-17359-2 PMID:32859912 PMCID: PMC7455744 |
 |
229.
Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. (pdf)
Journal: Nature Genetics, 2020 Mar doi: 10.1038/s41588-019-0562-0 PMID:32024998 PMCID: PMC7058536 |
 |
228.
Pathway and network analysis of more than 2500 whole cancer genomes. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-020-14367-0 PMID:32024854 PMCID: PMC7002574 |
 |
226.
Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. (pdf)
Journal: Communications Biology, 2020 Feb 5 doi: 10.1038/s42003-019-0741-7 PMID:32024996 PMCID: PMC7002399 |
 |
225.
Integrative pathway enrichment analysis of multivariate omics data. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-019-13983-9 PMID:32024846 PMCID: PMC7002665 |
 |
221.
Combined burden and functional impact tests for cancer driver discovery using DriverPower. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-019-13929-1 PMID:32024818 PMCID: PMC7002750 |
 |
216.
Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. (pdf)
Journal: Nature, 2020 Feb doi: 10.1038/s41586-020-1965-x PMID:32025015 PMCID: PMC7054214 |
 |
205.
A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS). (pdf)
Journal: Nature Communications, 2019 Jun 28 doi: 10.1038/s41467-019-10912-8 PMID:31253799 PMCID: PMC6599391 |
 |
198.
An AR-ERG transcriptional signature defined by long-range chromatin interactomes in prostate cancer cells. (pdf)
Journal: Genome Research, 2019 Feb doi: 10.1101/gr.230243.117 PMID:30606742 PMCID: PMC6360806 |
 |
197.
ncdDetect2: improved models of the site-specific mutation rate in cancer and driver detection with robust significance evaluation. (pdf)
Journal: Bioinformatics, 2019 Jan 15 doi: 10.1093/bioinformatics/bty511 PMID:29945188 PMCID: PMC6330011 |
 |
194.
Loss of LDAH associated with prostate cancer and hearing loss. (pdf)
Journal: Human Molecular Genetics, 2018 Dec 15 doi: 10.1093/hmg/ddy310 PMID:30169630 PMCID: PMC6276829 |
 |
179.
Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. (pdf)
Journal: Npj Genomic Medicine, 2018 doi: 10.1038/s41525-017-0040-5 PMID:29354286 PMCID: PMC5765157 |
 |
168.
Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. (pdf)
Journal: Nature Genetics, 2017 Jun doi: 10.1038/ng.3841 PMID:28436984 PMCID: PMC5841952 |
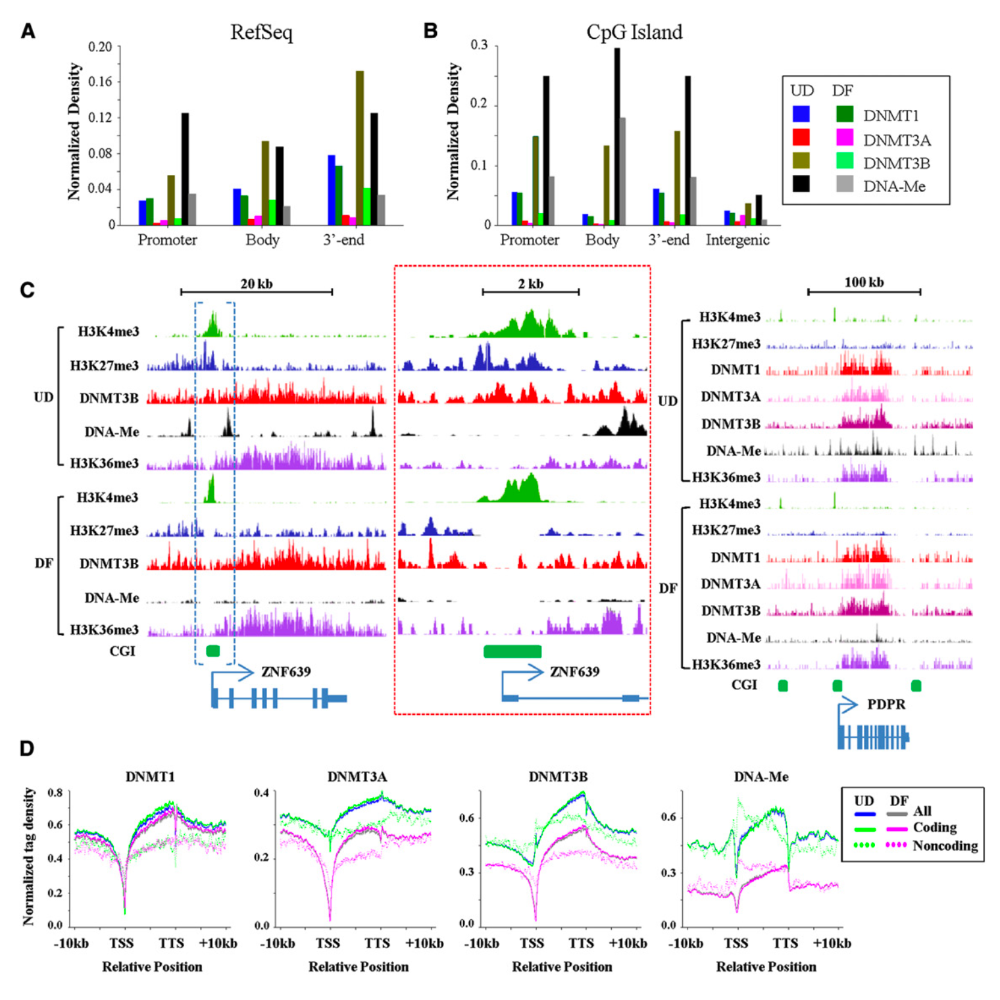 |
81.
Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. (pdf)
Journal: Cell Reports, 2012 Nov 29 doi: 10.1016/j.celrep.2012.10.017 PMID:23177624 PMCID: PMC3625945 |
 |
37.
The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. (pdf)
Journal: Science, 2010 Jan 1 doi: 10.1126/science.1180616 PMID:20044575 PMCID: PMC2982769 |
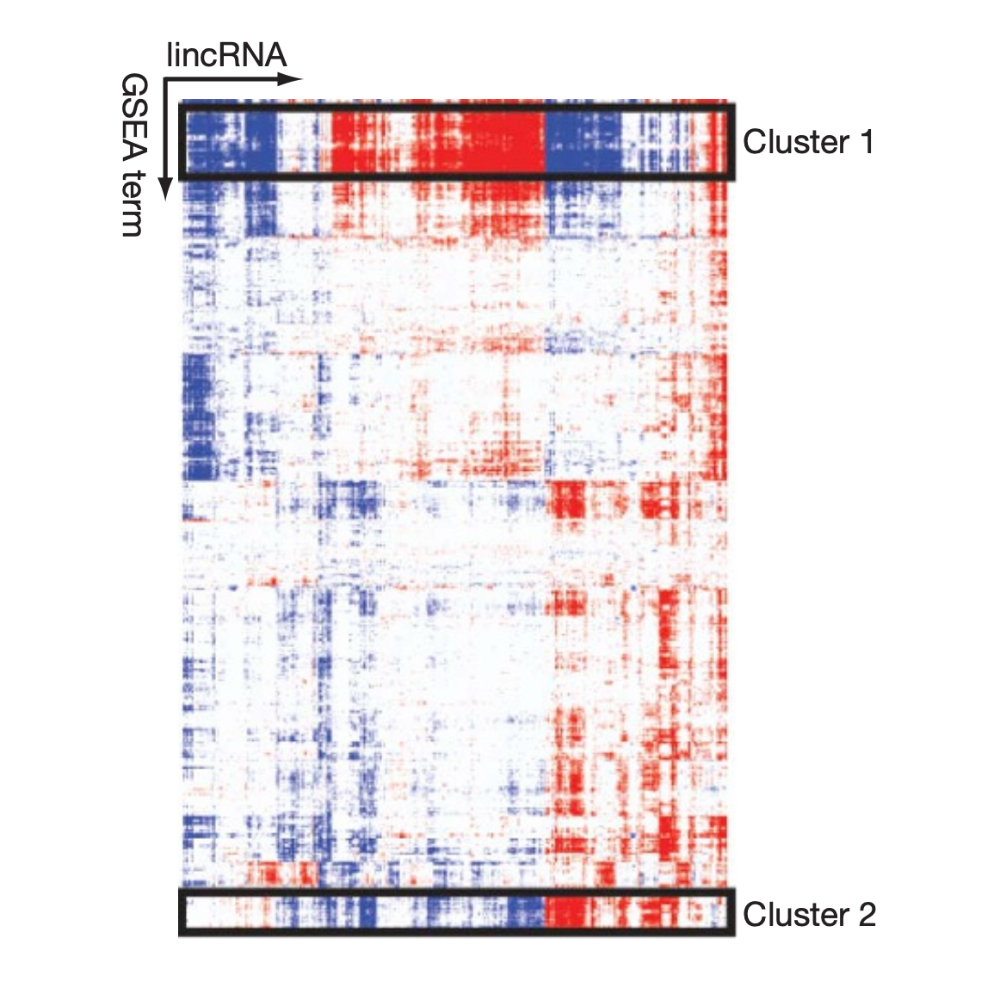
RNA Biology / Long Non-Coding RNAs / RNA Modifications
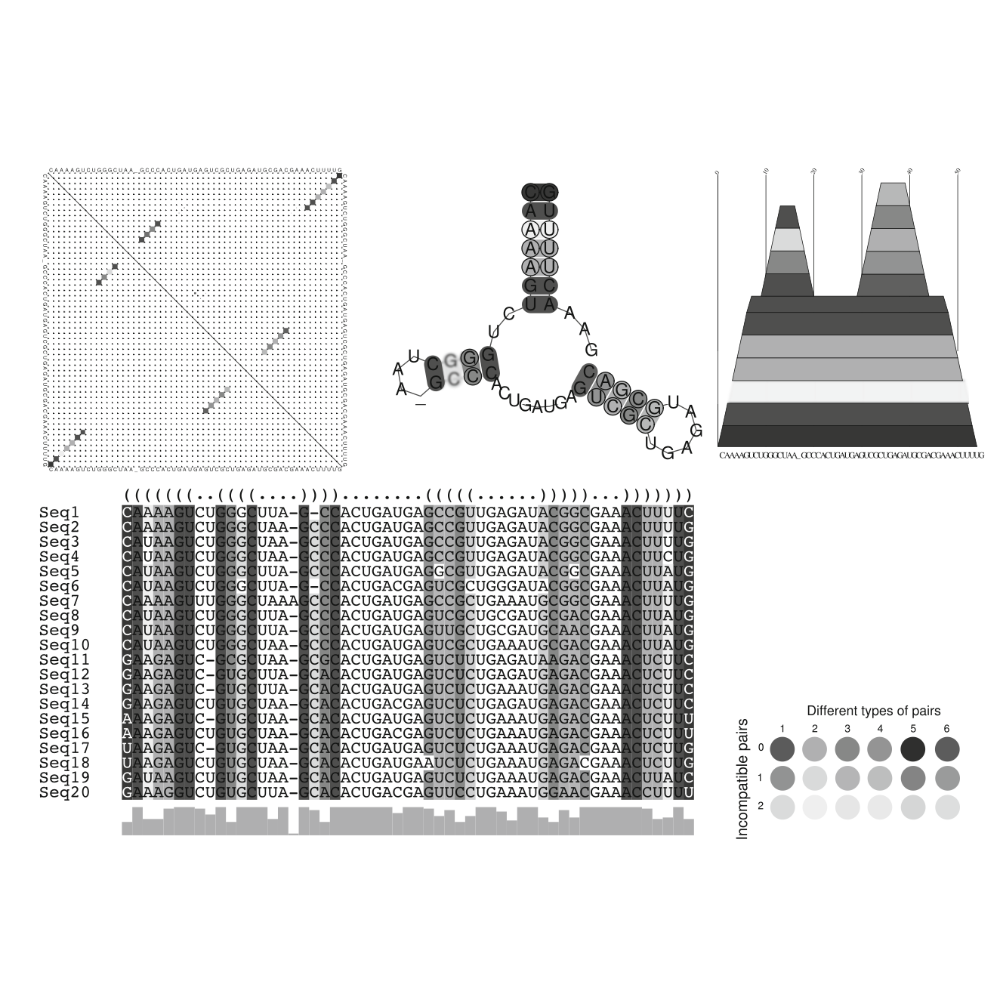
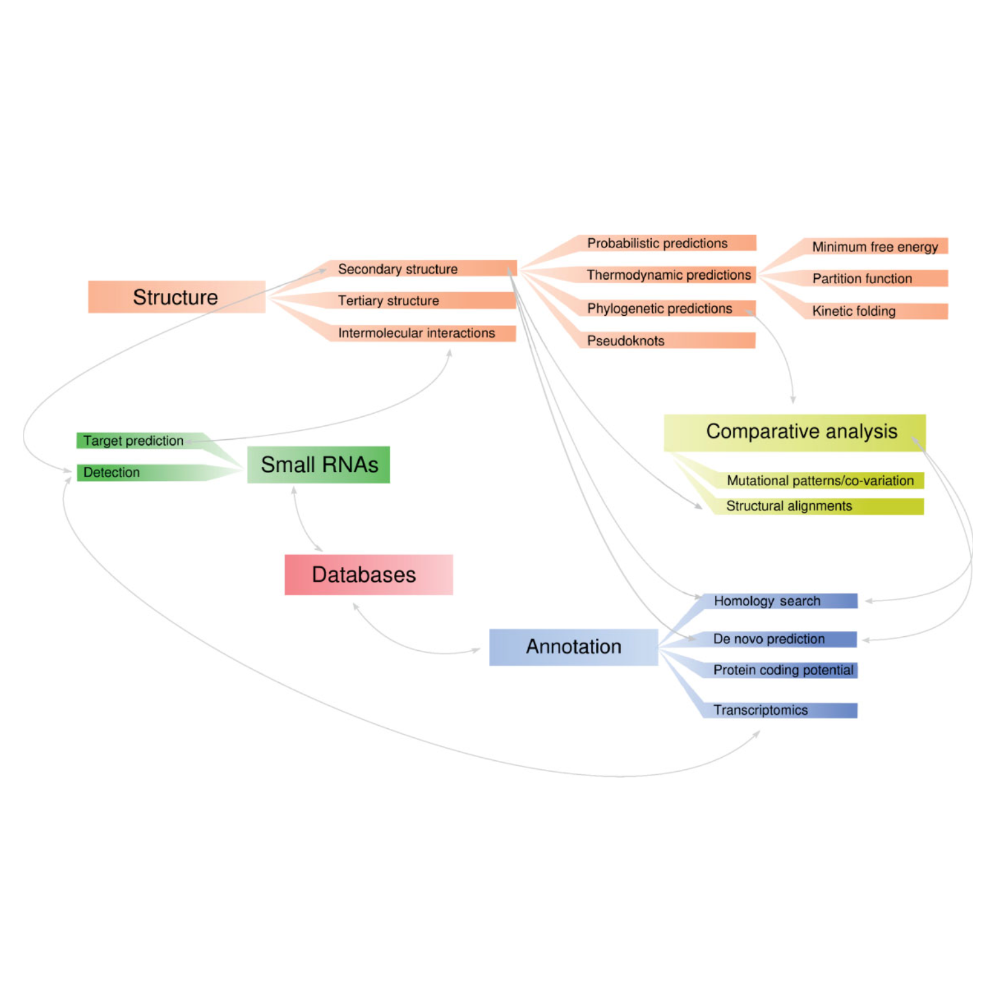
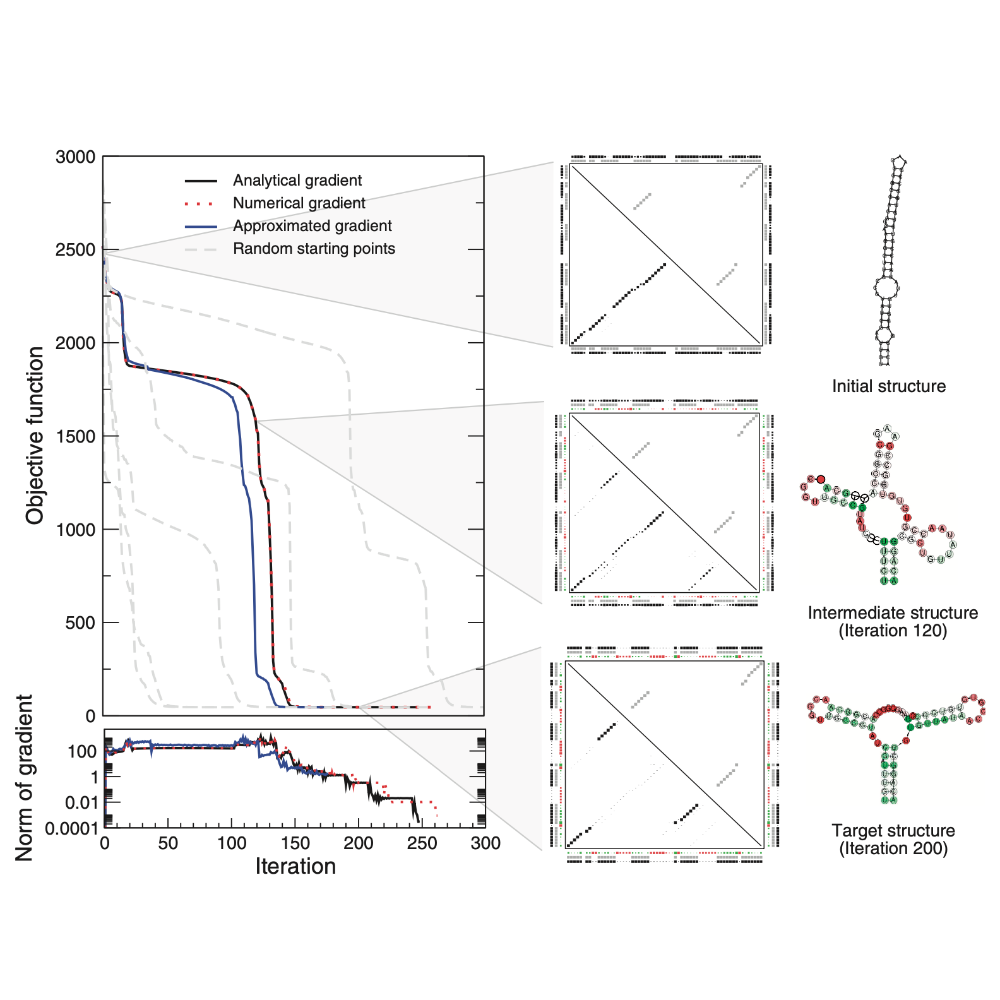
The recent realization that many long transcripts in the human genome do not encode proteins has opened up a whole new field for the study of long non-coding RNAs (lncRNAs). Two of our genomic signatures have facilitated their discovery and characterization, namely the chromatin signatures associated with promoters and transcribed regions, and the evolutionary signatures associated with protein-coding selection. This has enabled us to participate in numerous collaborations seeking the discovery, annotation, and functional characterization of long non-coding RNAs. We have also developed computational methods for studying the structural properties of non-coding RNAs, based on their evolutionary signatures, biophysical folding properties, and recent types of experimental evidence that distinguished paired vs. unpaired positions and can thus constrain the folding algorithms.
 |
226.
Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. (pdf)
Journal: Communications Biology, 2020 Feb 5 doi: 10.1038/s42003-019-0741-7 PMID:32024996 PMCID: PMC7002399 |
 |
200.
A gene expression atlas of embryonic neurogenesis in Drosophila reveals complex spatiotemporal regulation of lncRNAs. (pdf)
Journal: Development, 2019 Mar 28 doi: 10.1242/dev.175265 PMID:30923056 PMCID: PMC6451322 |
 |
180.
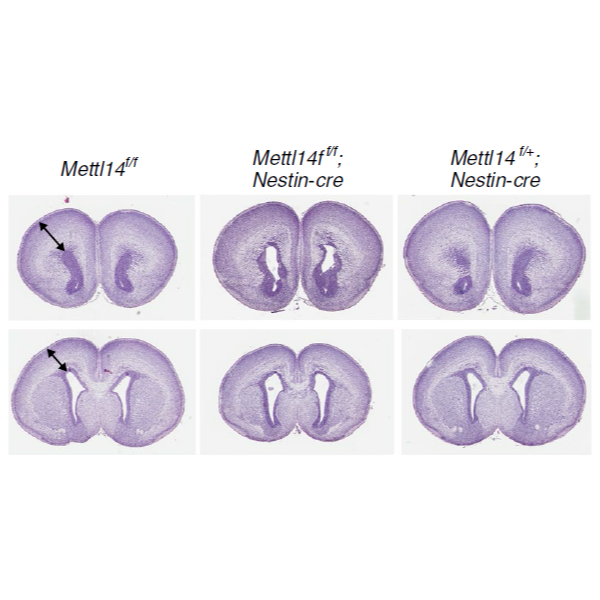
N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. (pdf)
Journal: Nature Neuroscience, 2018 Feb doi: 10.1038/s41593-017-0057-1 PMID:29335608 PMCID: PMC6317335 |
 |
172.
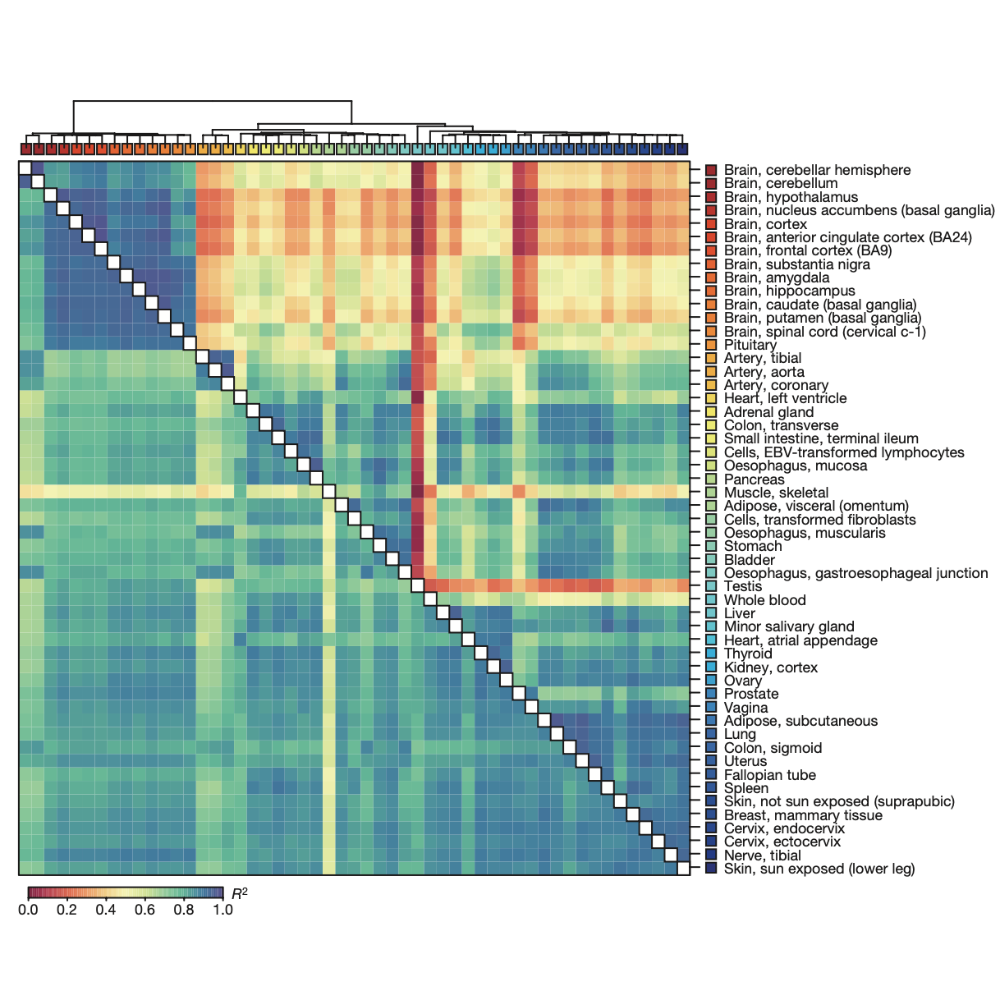
Dynamic landscape and regulation of RNA editing in mammals. (pdf)
Journal: Nature, 2017 Oct 11 doi: 10.1038/nature24041 PMID:29022589 PMCID: PMC5723435 |
 |
163.
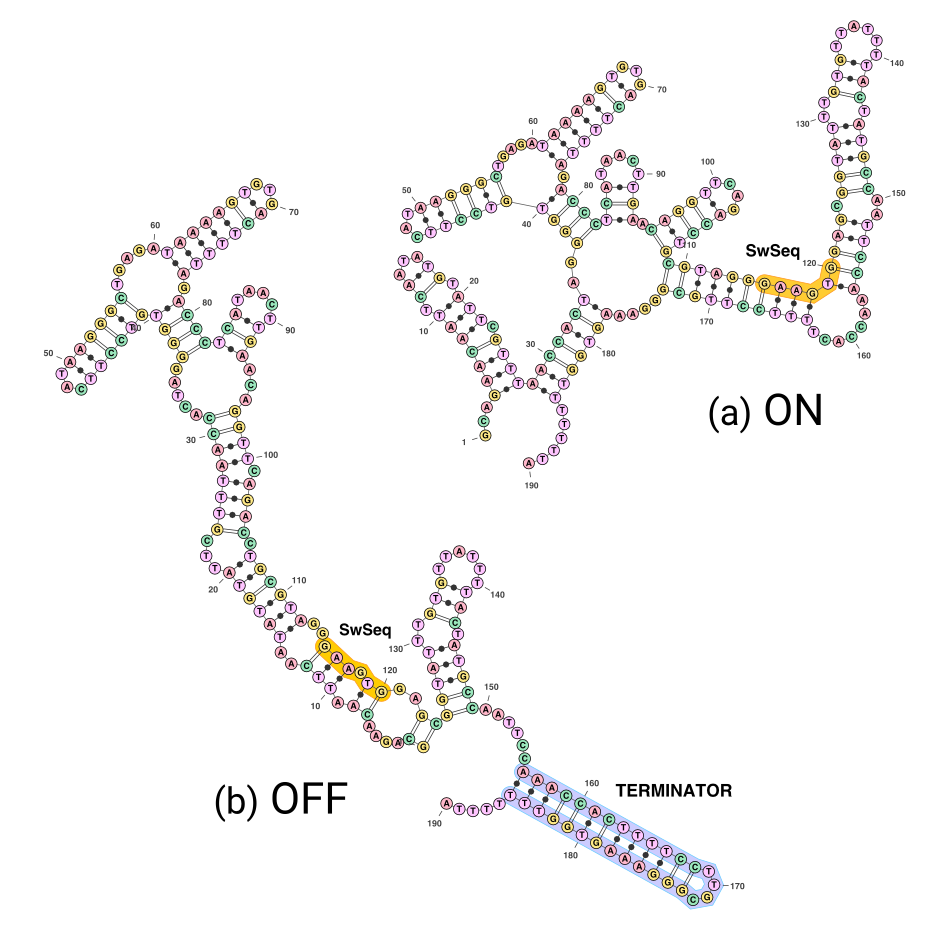
SwiSpot: modeling riboswitches by spotting out switching sequences. (pdf)
Journal: Bioinformatics, 2016 Nov 1 doi: 10.1093/bioinformatics/btw401 PMID:27378291 |
 |
111.
Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. (pdf)
Journal: Genome Research, 2014 Apr doi: 10.1101/gr.165035.113 PMID:24429298 PMCID: PMC3975061 |
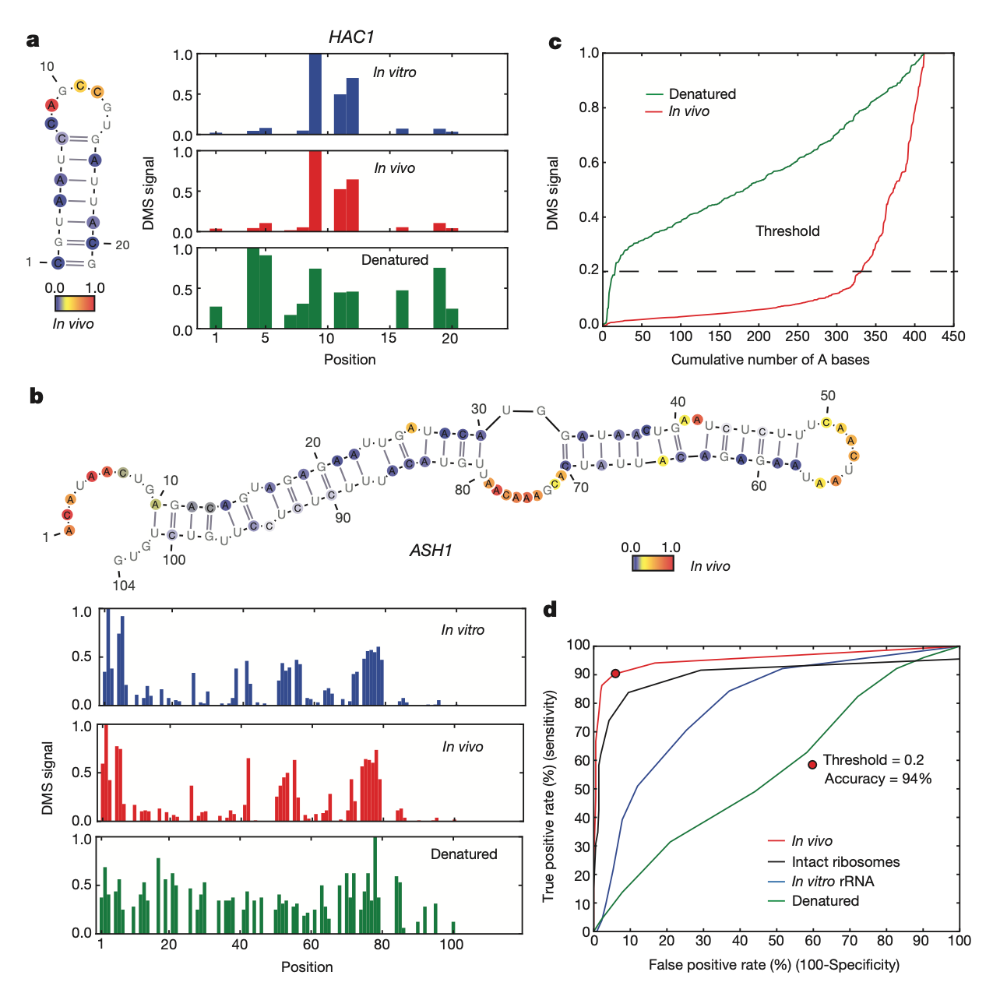 |
106.
Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. (pdf)
Journal: Nature, 2014 Jan 30 doi: 10.1038/nature12894 PMID:24336214 PMCID: PMC3966492 |
 |
105.
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments. (pdf)
Journal: Methods In Molecular Biology, 2014 doi: 10.1007/978-1-62703-709-9_7 PMID:24639158 |
 |
104.
Multiple knockout mouse models reveal lincRNAs are required for life and brain development. (pdf)
Journal: Elife, 2013 Dec 31 doi: 10.7554/eLife.01749 PMID:24381249 PMCID: PMC3874104 |
 |
89.
Long noncoding RNAs regulate adipogenesis. (pdf)
Journal: Proceedings Of The National Academy Of Sciences Of The United States Of America, 2013 Feb 26 doi: 10.1073/pnas.1222643110 PMID:23401553 PMCID: PMC3587215 |
 |
87.
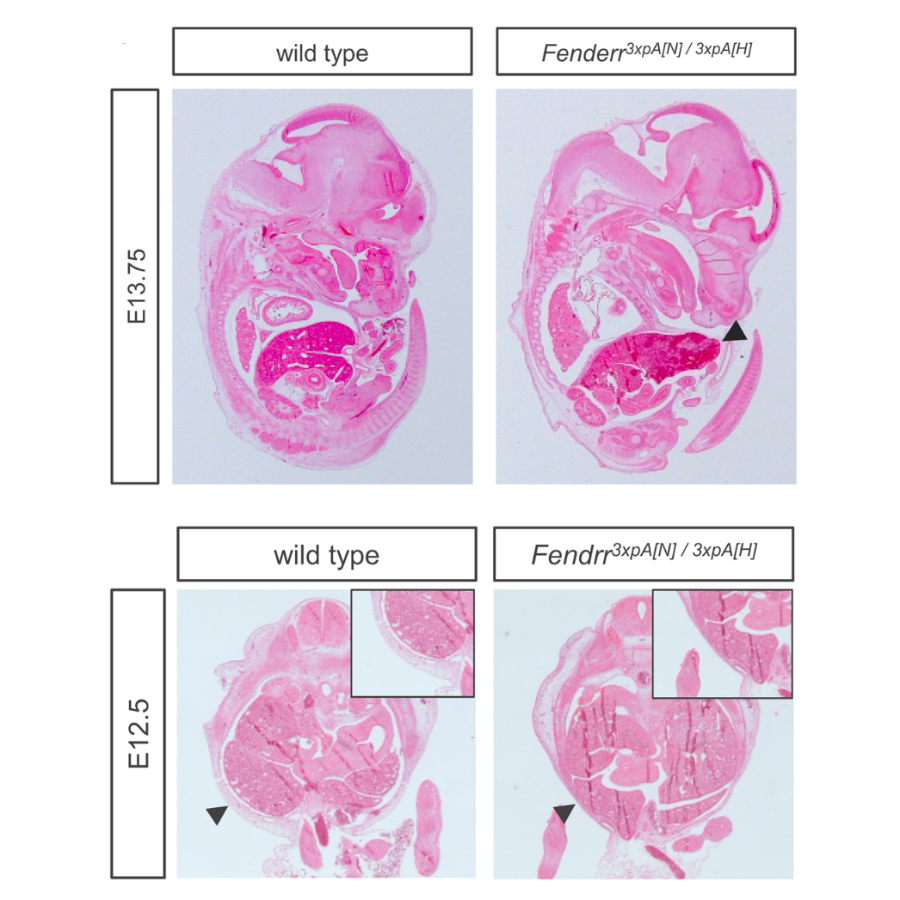
The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. (pdf)
Journal: Developmental Cell, 2013 Jan 28 doi: 10.1016/j.devcel.2012.12.012 PMID:23369715 PMCID: PMC4149175 |
 |
82.
Disruption of a large intergenic noncoding RNA in subjects with neurodevelopmental disabilities. (pdf)
Journal: American Journal Of Human Genetics, 2012 Dec 7 doi: 10.1016/j.ajhg.2012.10.016 PMID:23217328 PMCID: PMC3516594 |
 |
79.
Computational analysis of noncoding RNAs. (pdf)
Journal: Wiley Interdisciplinary Reviews. Rna, 2012 Nov-Dec doi: 10.1002/wrna.1134 PMID:22991327 PMCID: PMC3472101 |
 |
69.
RNA folding with soft constraints: reconciliation of probing data and thermodynamic secondary structure prediction. (pdf)
Journal: Nucleic Acids Research, 2012 May doi: 10.1093/nar/gks009 PMID:22287623 PMCID: PMC3378861 |
 |
58.
New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. (pdf)
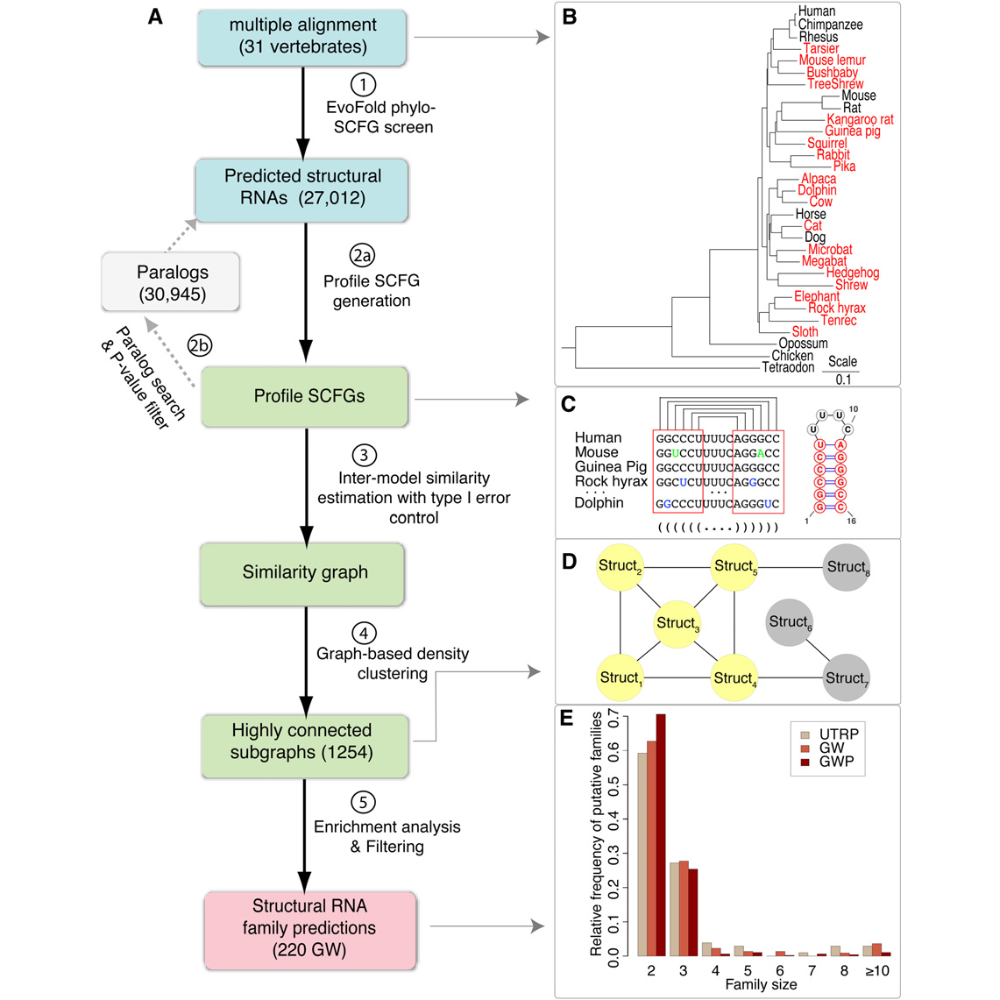
Journal: Genome Research, 2011 Nov doi: 10.1101/gr.112516.110 PMID:21994249 PMCID: PMC3205577 |
 |
53.
Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. (pdf)
Journal: Nature Genetics, 2011 Jun 5 doi: 10.1038/ng.848 PMID:21642992 PMCID: PMC3652667 |
 |
30.
Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. (pdf)
Journal: Nature, 2009 Mar 12 doi: 10.1038/nature07672 PMID:19182780 PMCID: PMC2754849 |
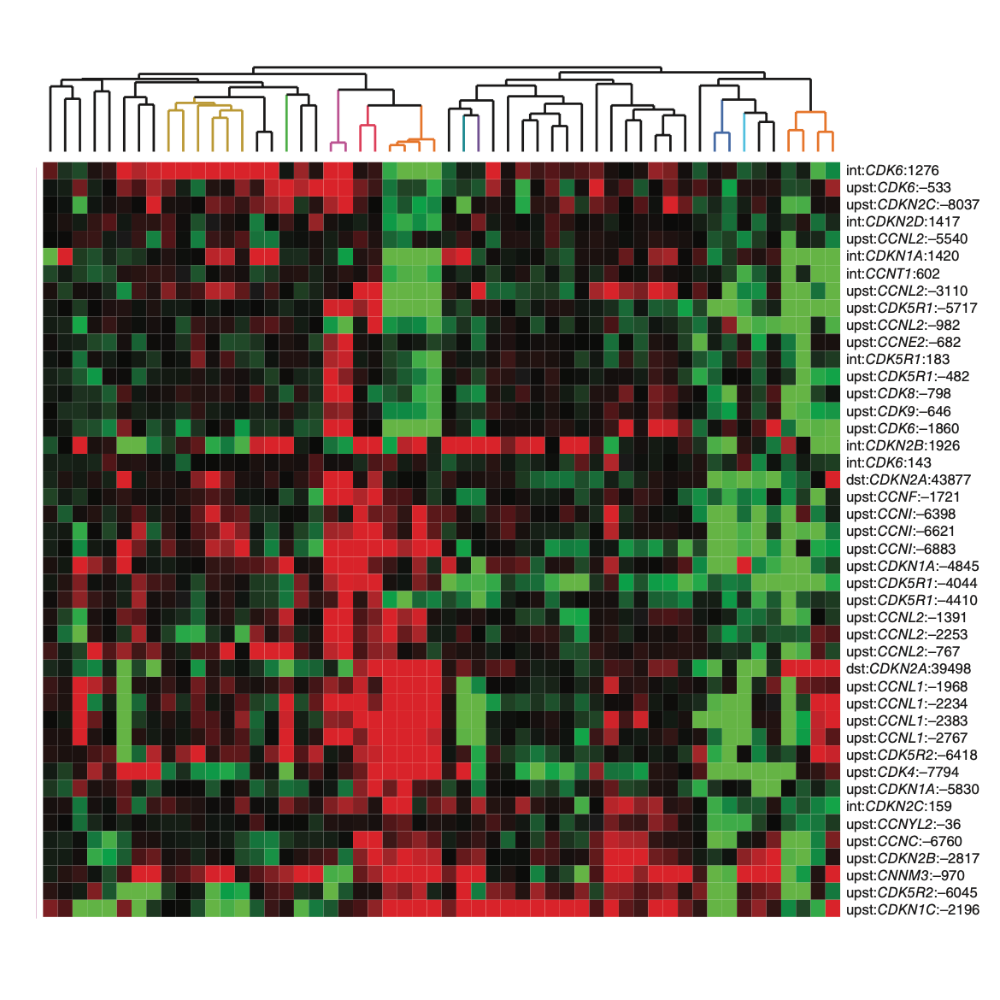
Small Regulatory RNAs
While gene regulation has been traditionally thought as mostly pre-transcriptional, it has been recently recognized that small regulatory RNAs can mediate the level of gene expression and translation post-transcriptionally, by directly targeting the messenger RNA of a protein-coding transcript. The most well-studied of these small regulatory RNAs are microRNAs (miRNAs), which recognize small 7-mer 'seed' motifs in the 3'untranslated regions (3'UTRs) of their targets. However, many additional classes of small regulatory RNAs have emerged, including piwiRNAs (piRNAs) encoded in a small number of master loci and specifically targeting and degrading transposable elements, and small-interfering RNAs (siRNAs) that can target both protein-coding and mobile elements and can arise from convergent transcripts or structured loci. Our group has developed methods for recognizing microRNAs based on their 3'UTR motif complementarity, their evolutionary signatures, and their sequencing signatures in RNA-Seq datasets. We have also developed methods for microRNA target prediction using evolutionary conservation. We have also shown that a single microRNA locus can result in two functional gene products by transcription of both strands into distinct and functional microRNAs. We have also participated in the discovery of piwiRNAs and endogenous siRNAs in Drosophila.
 |
165.
MicroRNA and gene expression changes in unruptured human cerebral aneurysms. (pdf)
Journal: Journal Of Neurosurgery, 2016 Dec doi: 10.3171/2015.11.JNS151841 PMID:26918470 PMCID: PMC5001931 |
 |
37.
The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. (pdf)
Journal: Science, 2010 Jan 1 doi: 10.1126/science.1180616 PMID:20044575 PMCID: PMC2982769 |
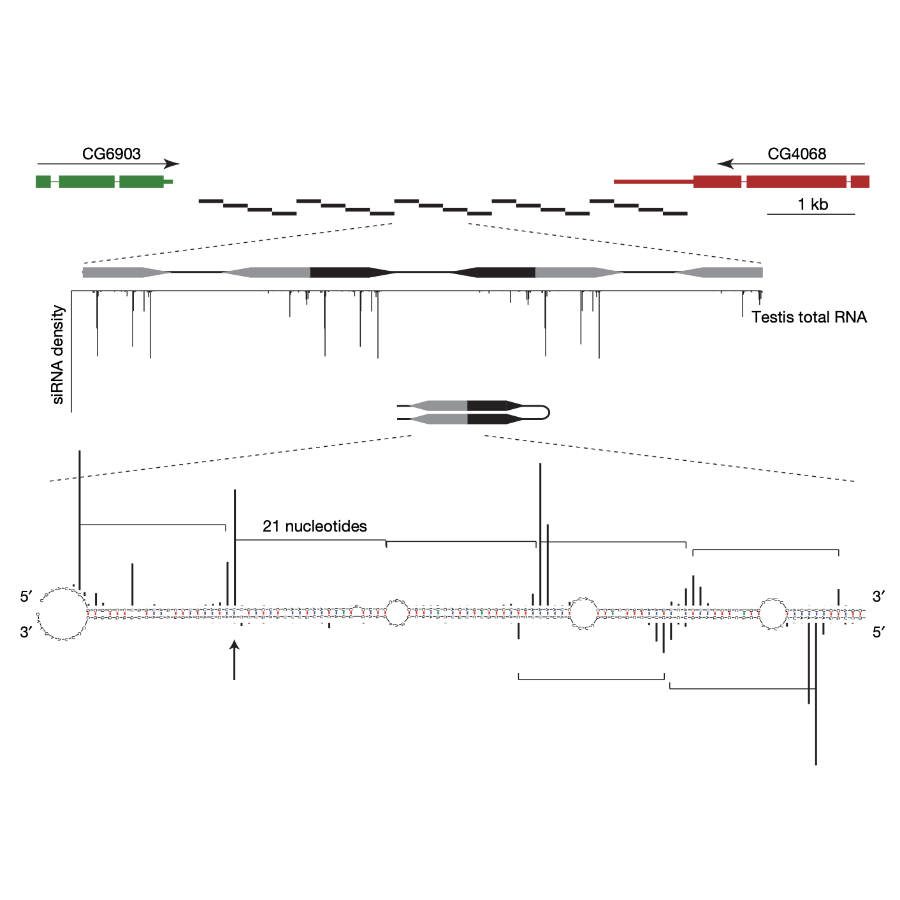 |
29.
An endogenous small interfering RNA pathway in Drosophila. (pdf)
Journal: Nature, 2008 Jun 5 doi: 10.1038/nature07007 PMID:18463631 PMCID: PMC2895258 |
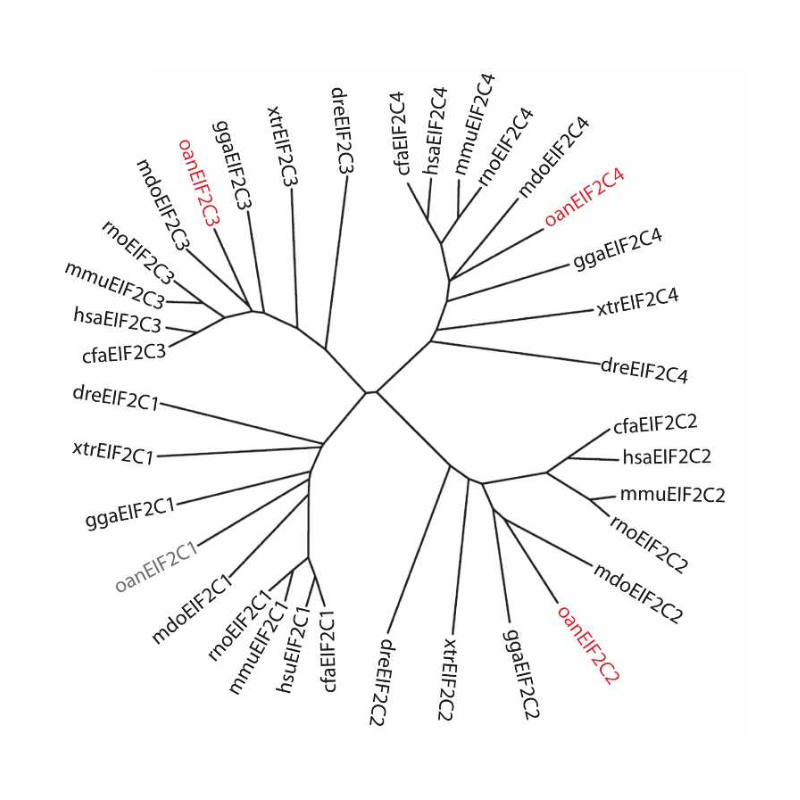 |
28.
Conservation of small RNA pathways in platypus. (pdf)
Journal: Genome Research, 2008 Jun doi: 10.1101/gr.073056.107 PMID:18463306 PMCID: PMC2413167 |
 |
24.
A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. (pdf)
Journal: Genes & Development, 2008 Jan 1 doi: 10.1101/gad.1613108 PMID:18172160 PMCID: PMC2151017 |
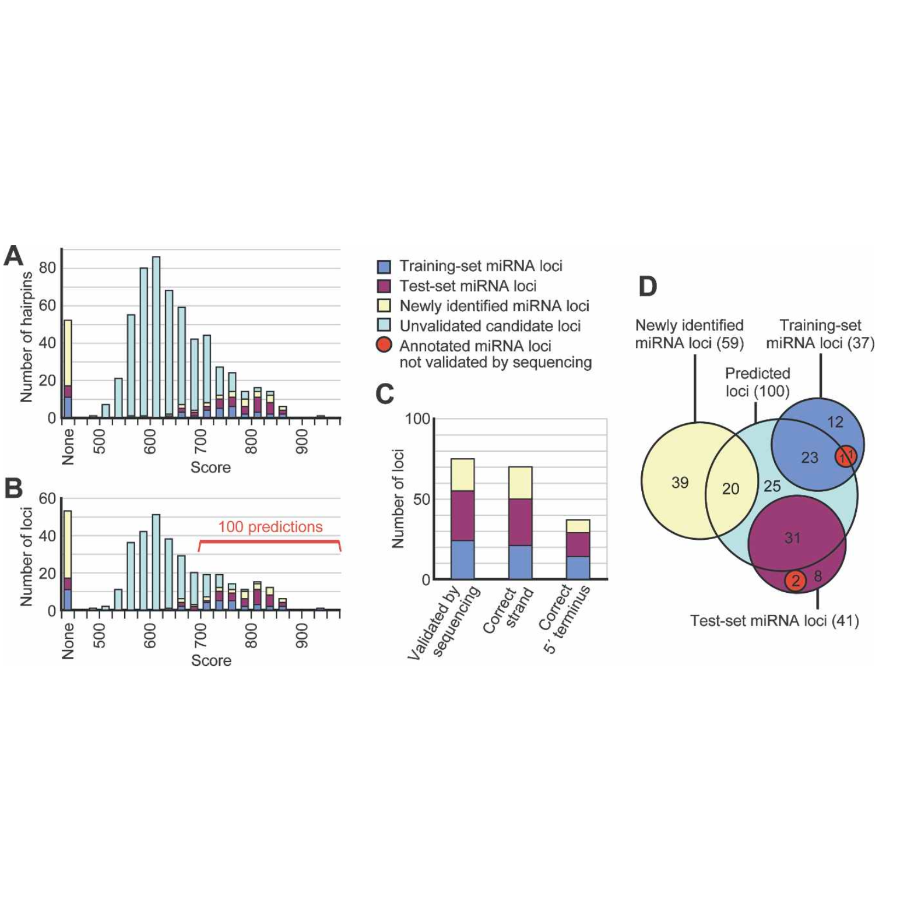 |
20.
Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. (pdf)
Journal: Genome Research, 2007 Dec doi: 10.1101/gr.6597907 PMID:17989254 PMCID: PMC2099593 |
 |
17.
Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. (pdf)
Journal: Genome Research, 2007 Dec doi: 10.1101/gr.6593807 PMID:17989255 PMCID: PMC2099594 |
 |
13.
Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. (pdf)
Journal: Cell, 2007 Mar 23 doi: 10.1016/j.cell.2007.01.043 PMID:17346786 |
 |
10.
Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. (pdf)
Journal: Nature, 2005 Mar 17 doi: 10.1038/nature03441 PMID:15735639 PMCID: PMC2923337 |
Protein-Coding Genes
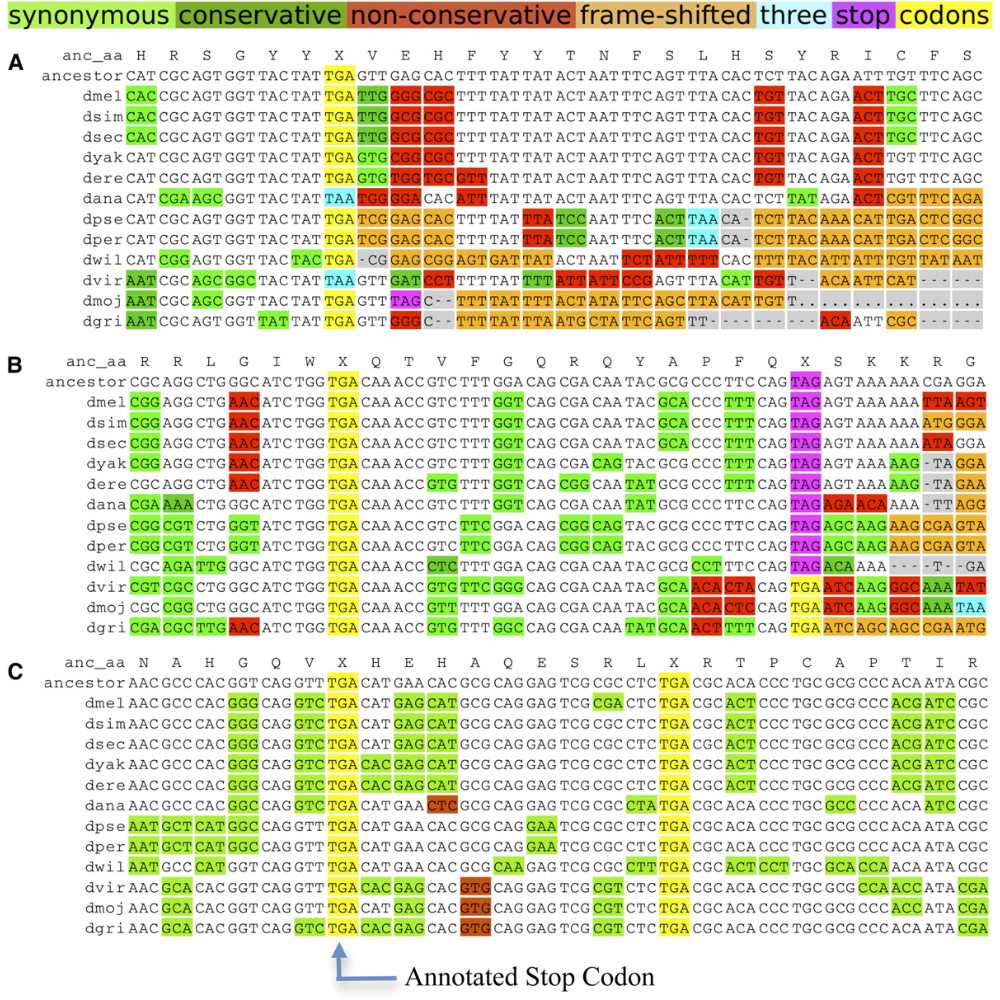
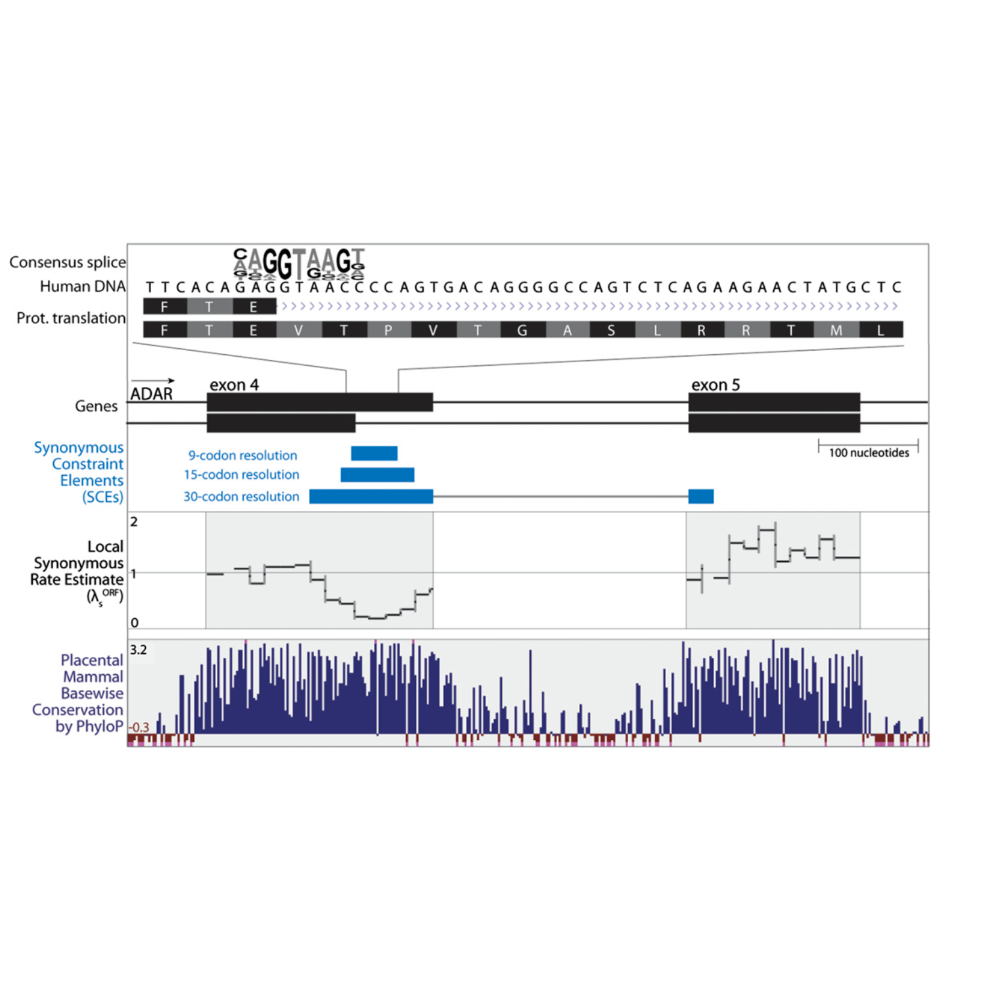
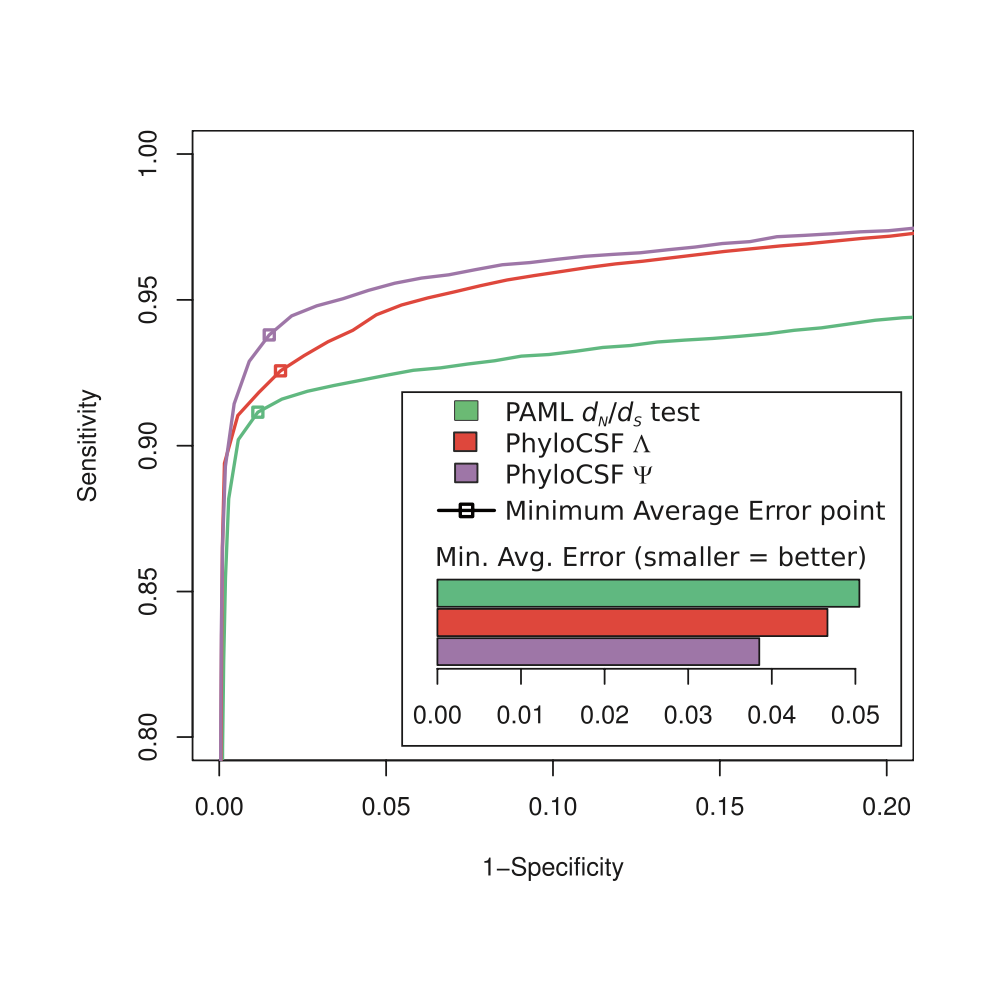
Protein-coding genes constitute the workhorse of the cell, and play almost every imaginable cellular role, from structural components to enzymes, regulators, signaling molecules, transporters. As such, human polymorphisms that disrupt protein-coding genes have been linked to countless diseases. Such studies rely on a systematic understanding of all the protein-coding genes encoded in the human genome. This has proven to be a daunting task, given the very short size of exons (only 50 nucleotides on average), the very long intergenic and intronic spaces (reaching hundreds of thousands of nucleotides), and the very complex patterns of alternative splicing that genes can undergo. To address this challenge, we have developed discriminative metrics for gene finding, based on comparative genomics evidence. We found several evolutionary signatures associated with protein-coding exons, including reading frame conservation (RFC) and codon substitution frequencies (CSF). We showed that these evolutionary signatures significantly outperform single-species and pairwise metrics, and applied them to revisit the yeast, fly, and human genomes, leading to new genes, new exons, and new alternative splice forms, revealing spurious annotations, refining gene boundaries and reading frame, and adding new alternative splice forms. We worked closely with curators and experimental labs, to validate most of these predictions, revising genome databases. These evolutionary signatures have also enabled us to reveal several types of unusual genes, that defy the traditional rules of translation. These include many polycistronic transcripts, genes with regulated stop codon readthrough events, and apparent programmed translational frameshifts, many of which correlate with mRNA secondary structures. Most of these results would have been inaccessible with traditional gene finding methods. We have also recently recognized that protein-coding exons can encode within them additional functional elements that exploit the degenerate nature of the genetic code.
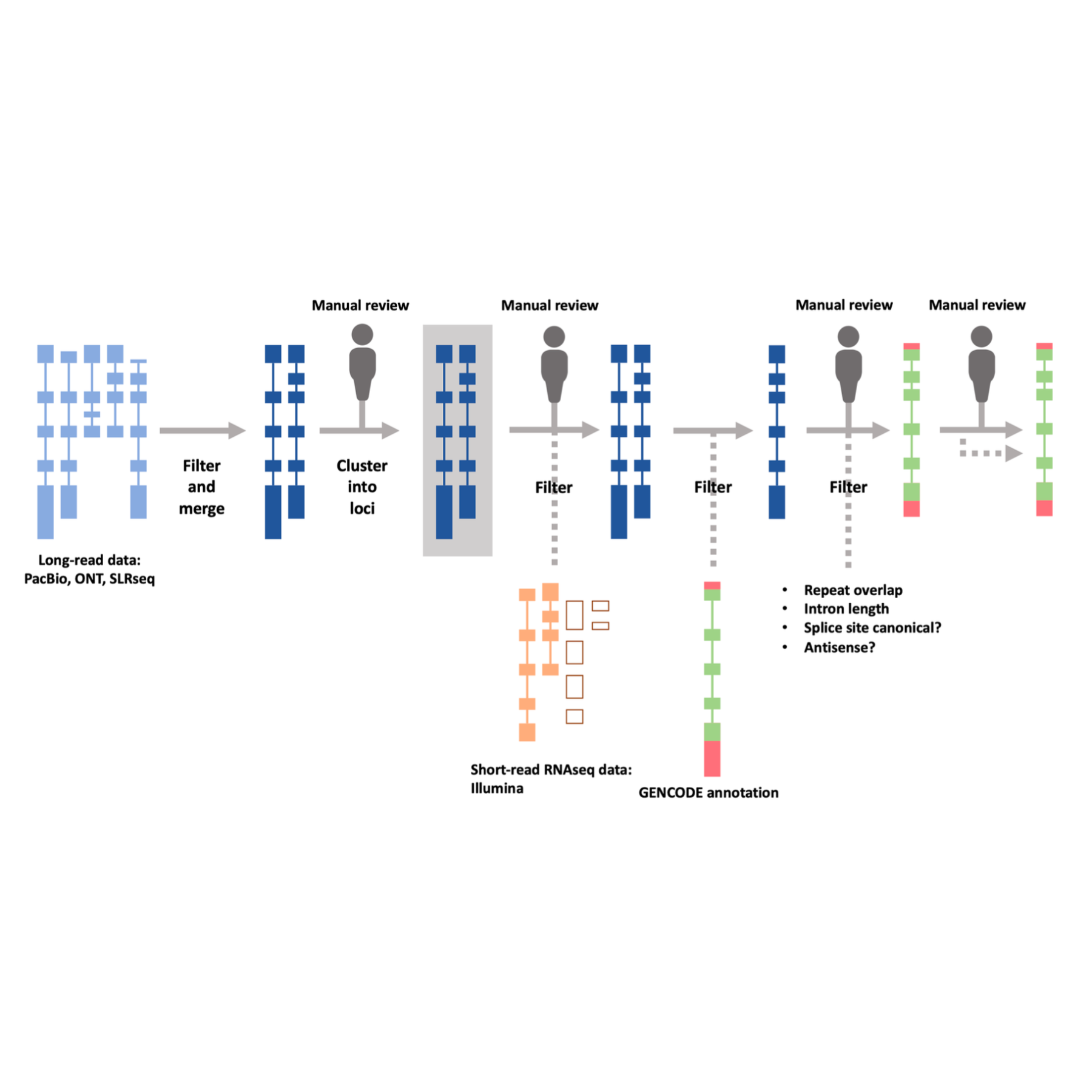 |
266.
GENCODE 2021. (pdf)
Journal: Nucleic Acids Research, 2021 Jan 8 doi: 10.1093/nar/gkaa1087 PMID:33270111 PMCID: PMC7778937 |
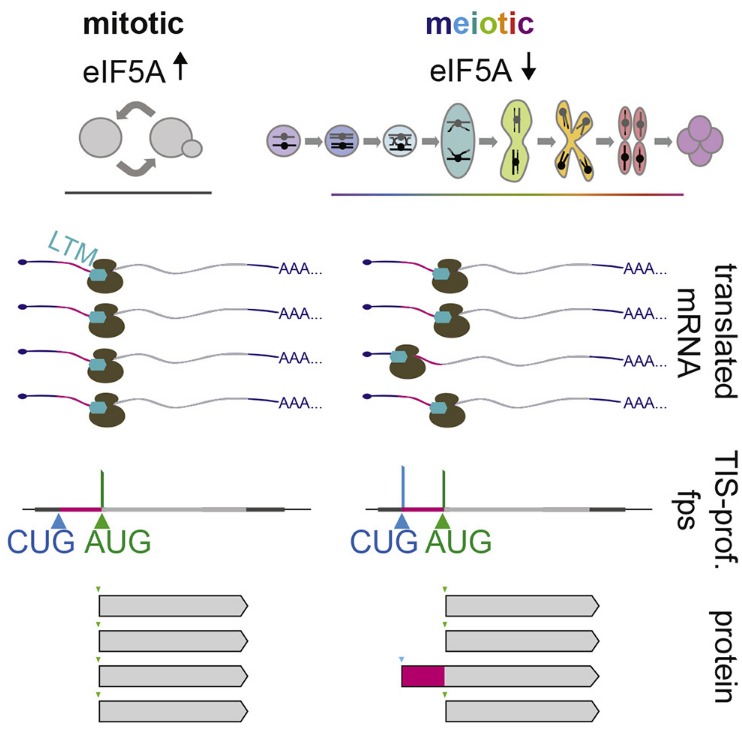 |
246.
Translation Initiation Site Profiling Reveals Widespread Synthesis of Non-AUG-Initiated Protein Isoforms in Yeast. (pdf)
Journal: Cell Systems, 2020 Aug 26 doi: 10.1016/j.cels.2020.06.011 PMID:32710835 PMCID: PMC7508262 |
 |
236.
Evidence for a novel overlapping coding sequence in POLG initiated at a CUG start codon. (pdf)
Journal: Bmc Genetics, 2020 Mar 6 doi: 10.1186/s12863-020-0828-7 PMID:32138667 PMCID: PMC7059407 |
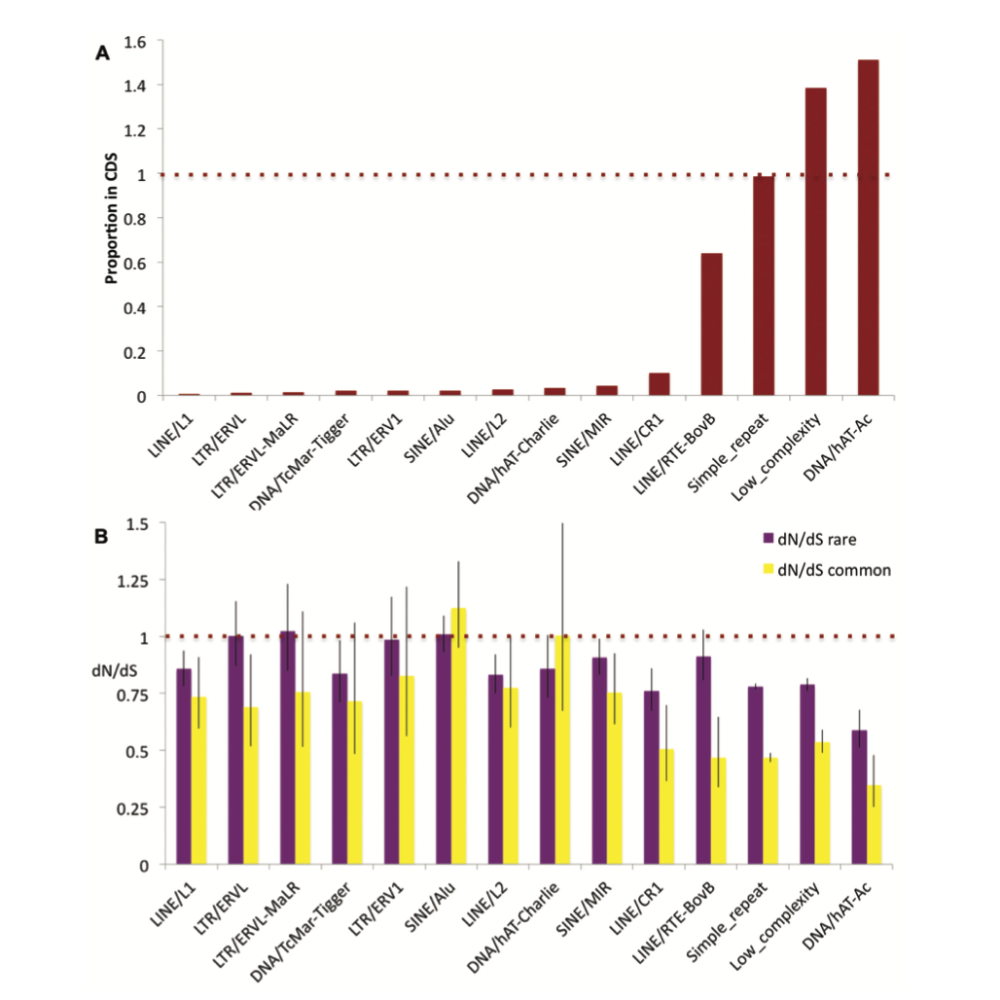 |
235.
Few SINEs of life: Alu elements have little evidence for biological relevance despite elevated translation. (pdf)
Journal: Nar Genomics And Bioinformatics, 2020 Mar doi: 10.1093/nargab/lqz023 PMID:31886458 PMCID: PMC6924539 |
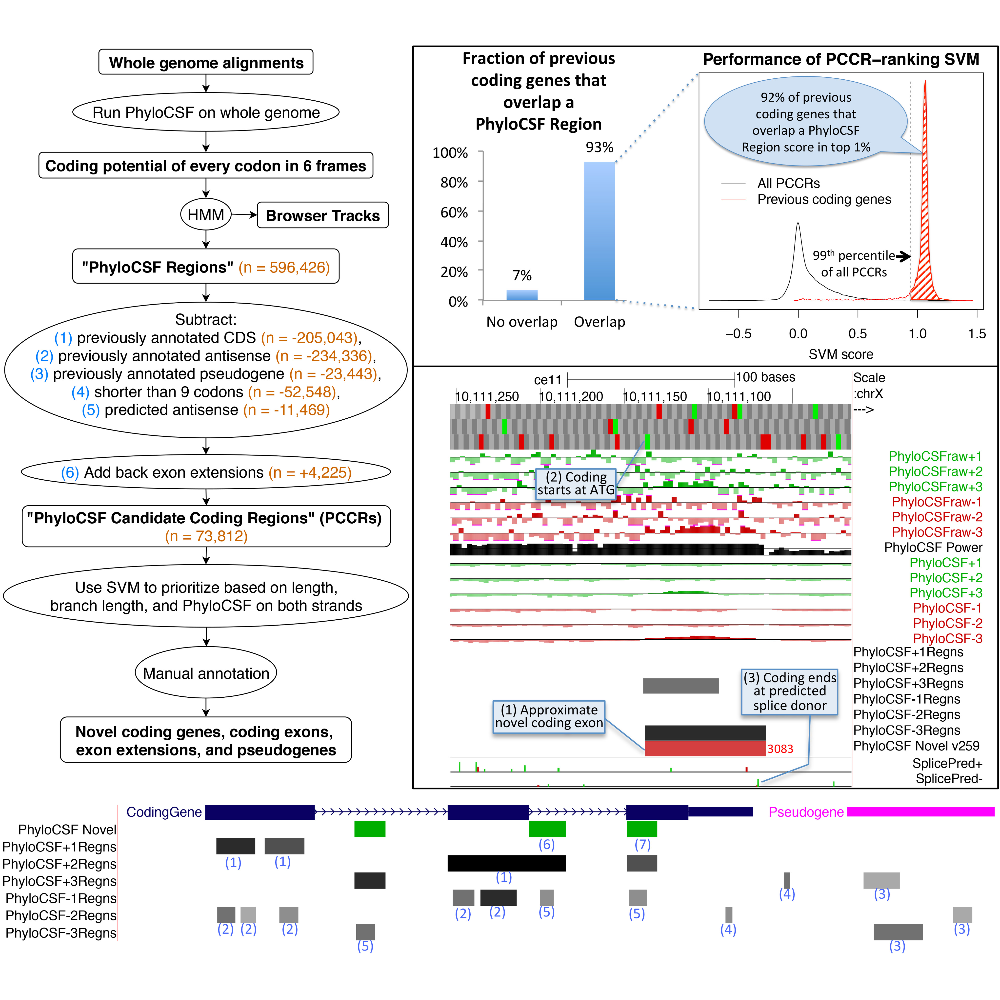 |
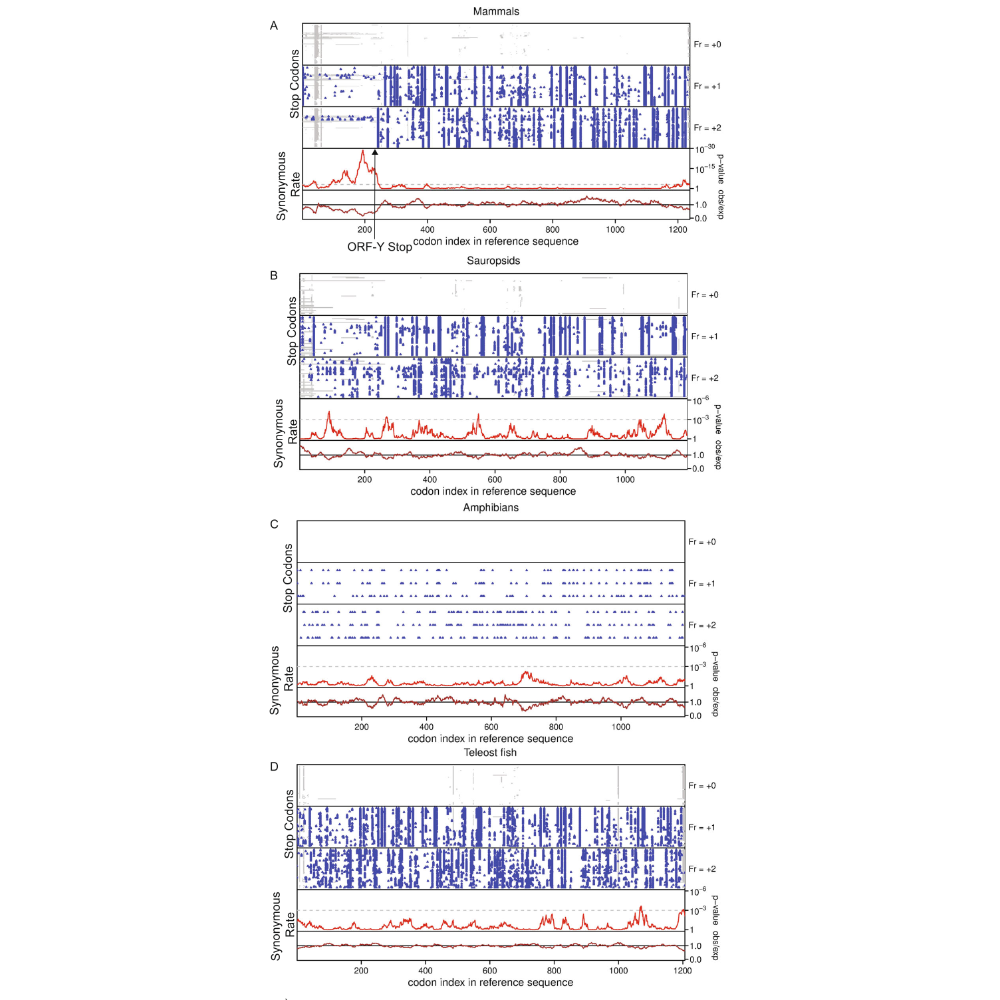
210.
Discovery of high-confidence human protein-coding genes and exons by whole-genome PhyloCSF helps elucidate 118 GWAS loci. (pdf)
Journal: Genome Research, 2019 Dec doi: 10.1101/gr.246462.118 PMID:31537640 PMCID: PMC6886504 |
 |
208.
Elucidation of Codon Usage Signatures across the Domains of Life. (pdf)
Journal: Molecular Biology And Evolution, 2019 Oct 1 doi: 10.1093/molbev/msz124 PMID:31220870 PMCID: PMC6759073 |
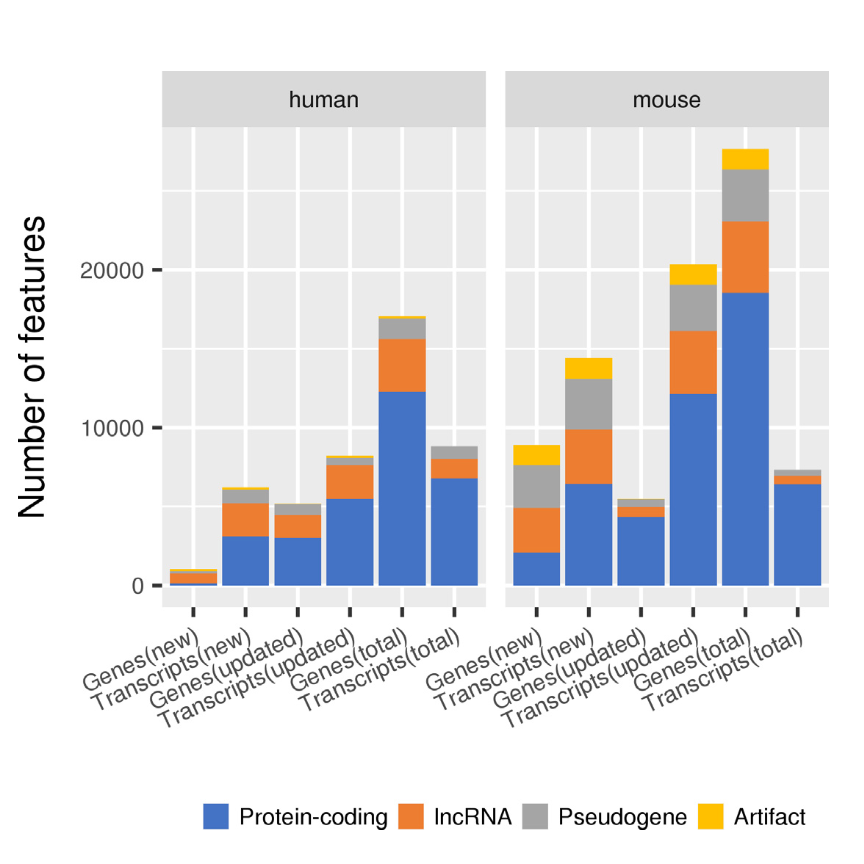 |
196.
GENCODE reference annotation for the human and mouse genomes. (pdf)
Journal: Nucleic Acids Research, 2019 Jan 8 doi: 10.1093/nar/gky955 PMID:30357393 PMCID: PMC6323946 |
 |
187.
Loose ends: almost one in five human genes still have unresolved coding status. (pdf)
Journal: Nucleic Acids Research, 2018 Aug 21 doi: 10.1093/nar/gky587 PMID:29982784 PMCID: PMC6101605 |
 |
186.
Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. (pdf)
Journal: Nature Structural & Molecular Biology, 2018 Aug doi: 10.1038/s41594-018-0091-z PMID:30061596 PMCID: PMC6690192 |
 |
181.
Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. (pdf)
Journal: The Journal Of Biological Chemistry, 2018 Mar 23 doi: 10.1074/jbc.M117.818526 PMID:29386352 PMCID: PMC5868278 |
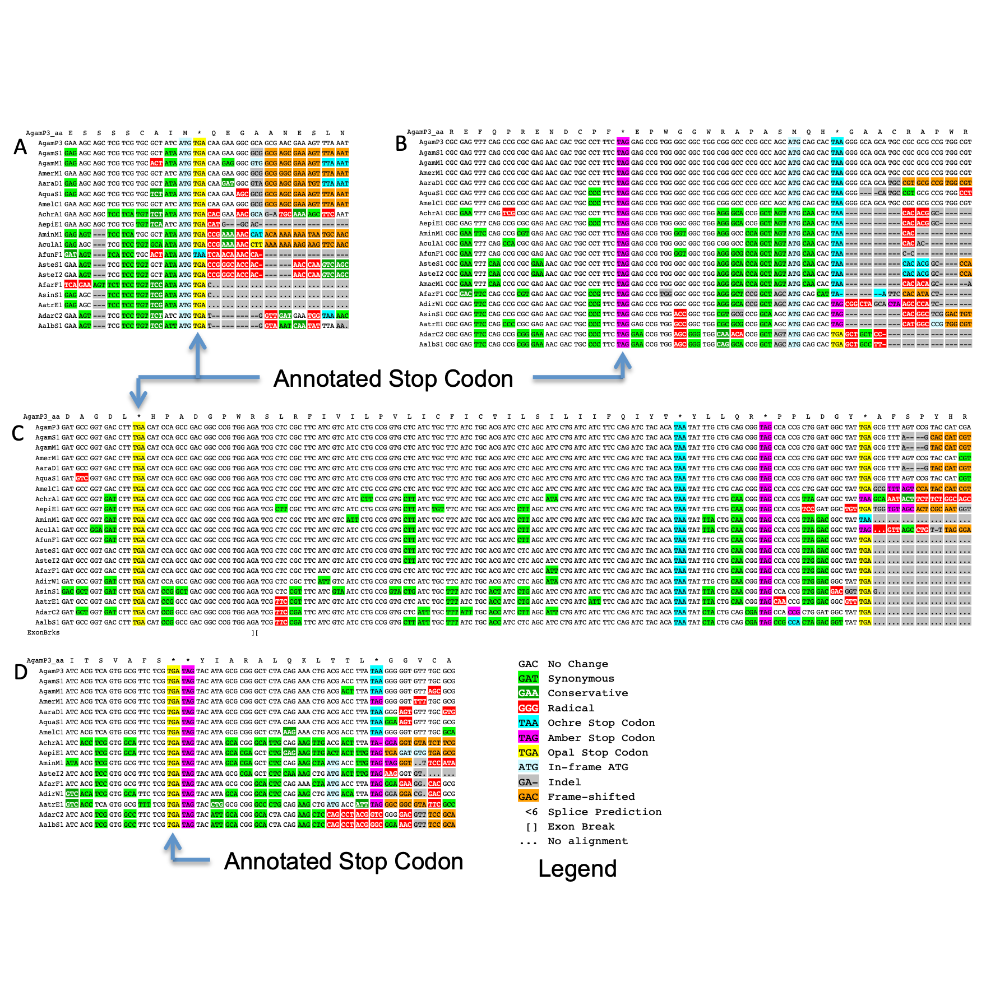 |
166.
Evolutionary Dynamics of Abundant Stop Codon Readthrough. (pdf)
Journal: Molecular Biology And Evolution, 2016 Dec doi: 10.1093/molbev/msw189 PMID:27604222 PMCID: PMC5100048 |
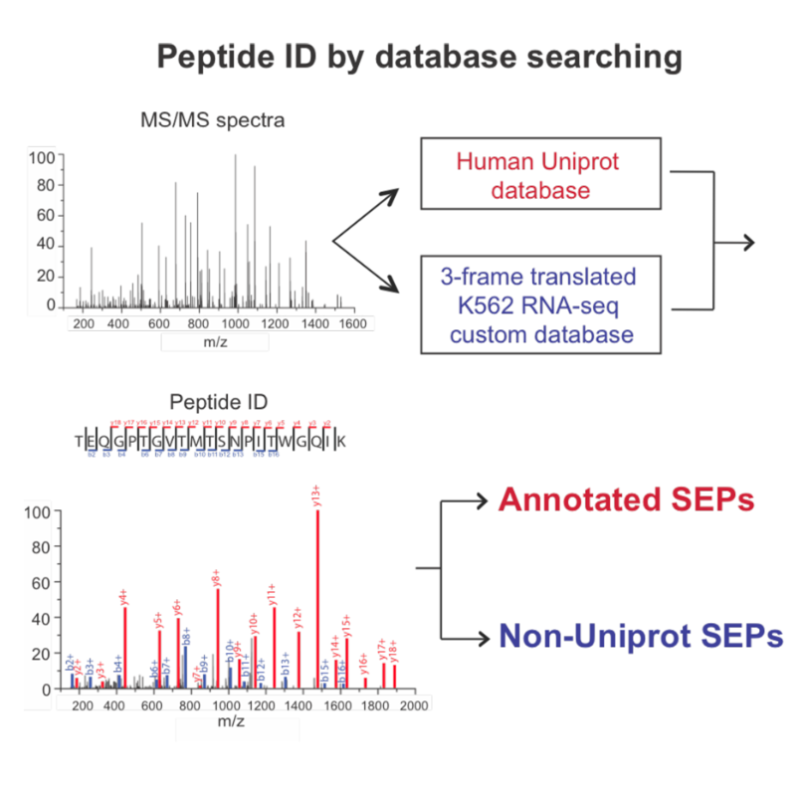 |
157.
Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. (pdf)
Journal: Analytical Chemistry, 2016 Apr 5 doi: 10.1021/acs.analchem.6b00191 PMID:27010111 PMCID: PMC4939623 |
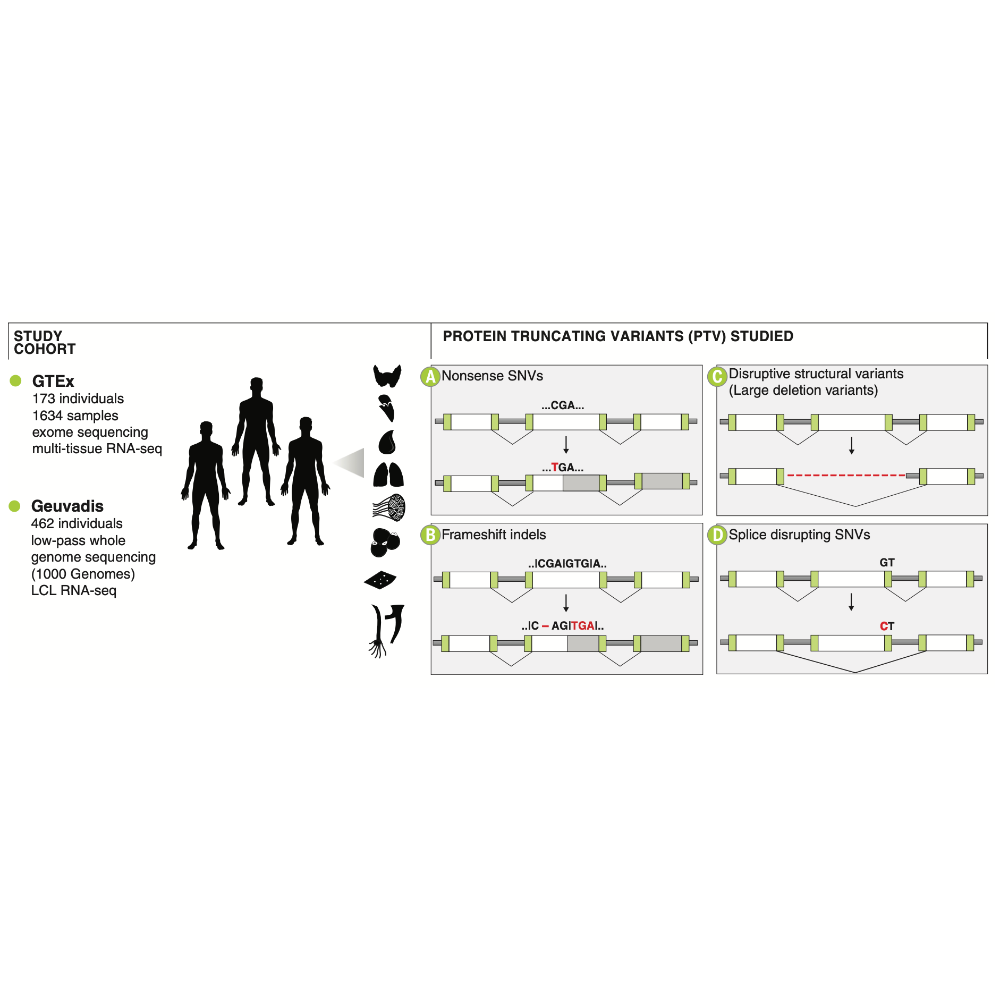 |
142.
Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. (pdf)
Journal: Science, 2015 May 8 doi: 10.1126/science.1261877 PMID:25954003 PMCID: PMC4537935 |
 |
130.
FRESCo: finding regions of excess synonymous constraint in diverse viruses. (pdf)
Journal: Genome Biology, 2015 Feb 17 doi: 10.1186/s13059-015-0603-7 PMID:25853568 PMCID: PMC4376164 |
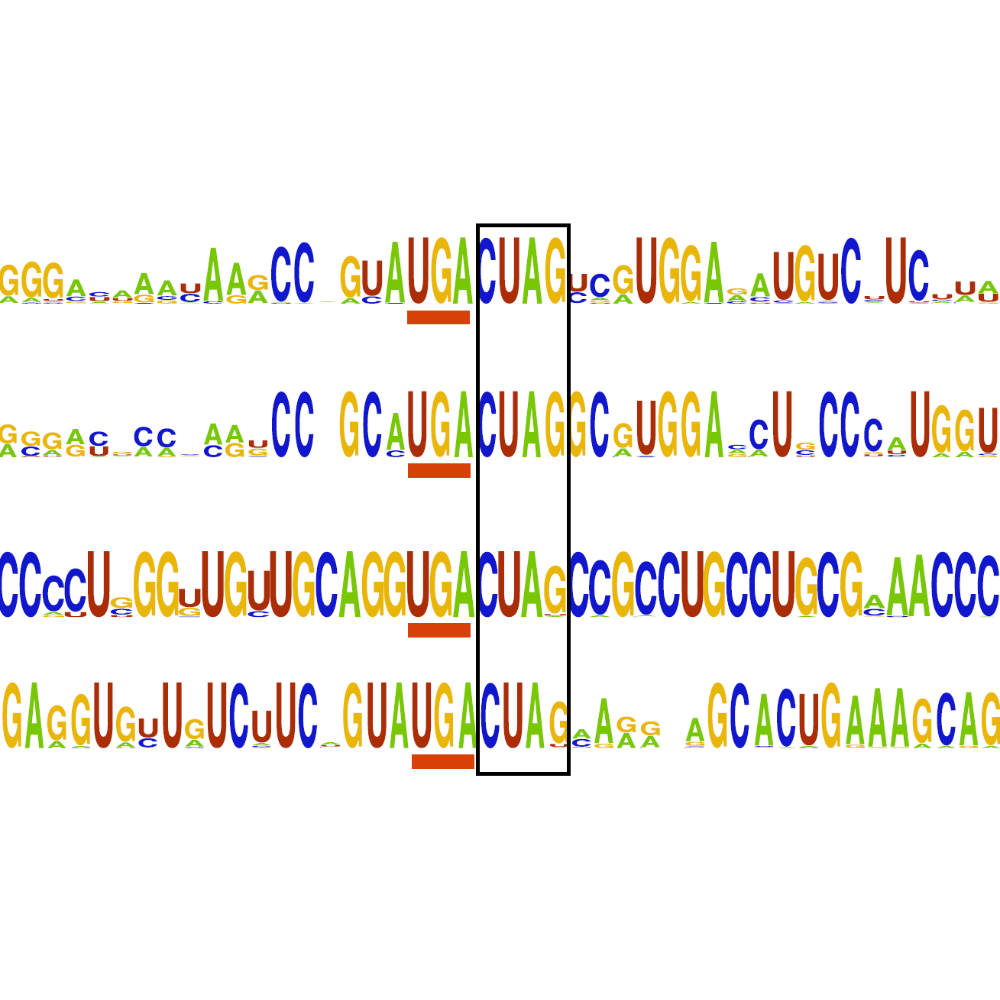 |
118.
Evidence of efficient stop codon readthrough in four mammalian genes. (pdf)
Journal: Nucleic Acids Research, 2014 Aug doi: 10.1093/nar/gku608 PMID:25013167 PMCID: PMC4132726 |
 |
109.
Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. (pdf)
Journal: Journal Of Proteome Research, 2014 Mar 7 doi: 10.1021/pr401280w PMID:24490786 PMCID: PMC3993966 |
 |
74.
GENCODE: the reference human genome annotation for The ENCODE Project. (pdf)
Journal: Genome Research, 2012 Sep doi: 10.1101/gr.135350.111 PMID:22955987 PMCID: PMC3431492 |
 |
60.
Evidence of abundant stop codon readthrough in Drosophila and other metazoa. (pdf)
Journal: Genome Research, 2011 Dec doi: 10.1101/gr.119974.110 PMID:21994247 PMCID: PMC3227100 |
 |
59.
Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. (pdf)
Journal: Genome Research, 2011 Nov doi: 10.1101/gr.108753.110 PMID:21994248 PMCID: PMC3205576 |
 |
54.
PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. (pdf)
Journal: Bioinformatics, 2011 Jul 1 doi: 10.1093/bioinformatics/btr209 PMID:21685081 PMCID: PMC3117341 |
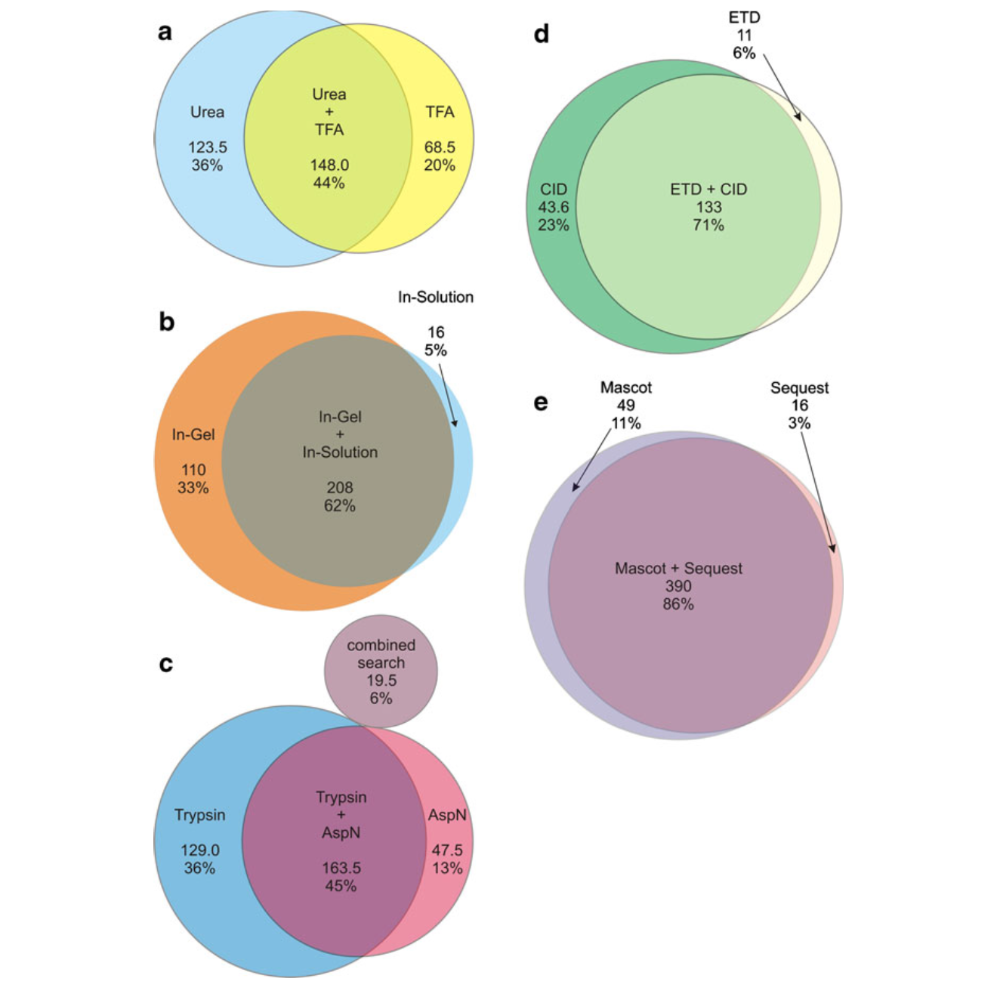 |
41.
Optimization of parameters for coverage of low molecular weight proteins. (pdf)
Journal: Analytical And Bioanalytical Chemistry, 2010 Dec doi: 10.1007/s00216-010-4093-x PMID:20803007 PMCID: PMC2990009 |
 |
34.
The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. (pdf)
Journal: Genome Research, 2009 Jul doi: 10.1101/gr.080531.108 PMID:19498102 PMCID: PMC2704439 |
 |
26.
Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. (pdf)
Journal: Plos Computational Biology, 2008 Apr 18 doi: 10.1371/journal.pcbi.1000067 PMID:18421375 PMCID: PMC2291194 |
 |
23.
Distinguishing protein-coding and noncoding genes in the human genome. (pdf)
Journal: Proceedings Of The National Academy Of Sciences Of The United States Of America, 2007 Dec 4 doi: 10.1073/pnas.0709013104 PMID:18040051 PMCID: PMC2148306 |
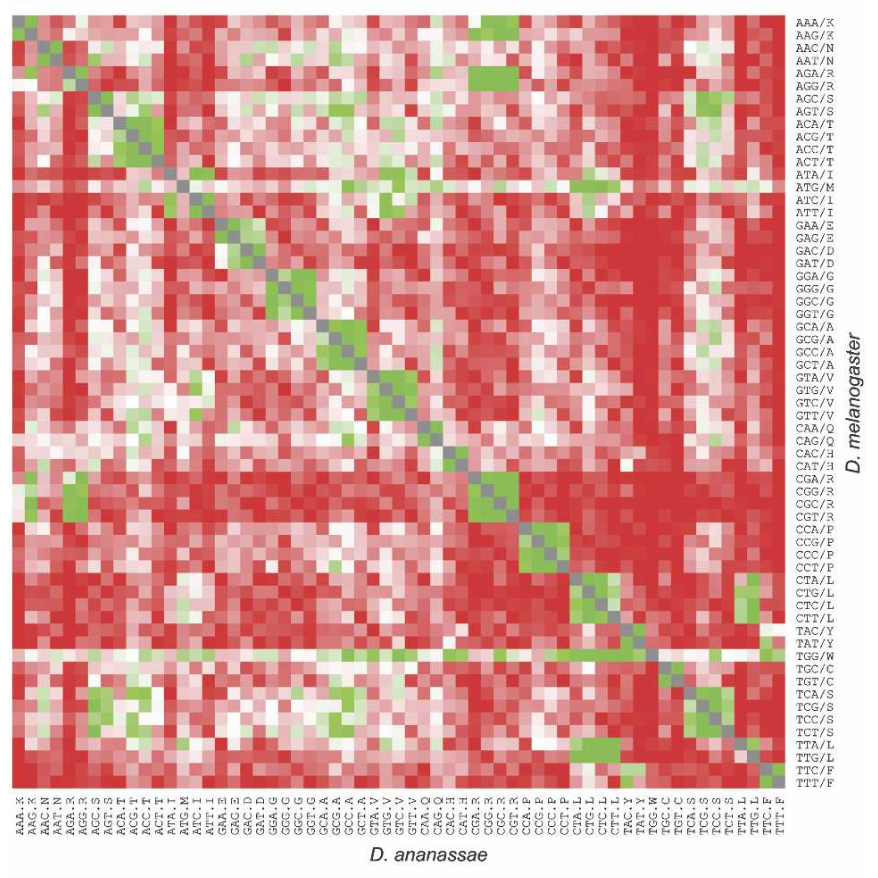 |
19.
Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. (pdf)
Journal: Genome Research, 2007 Dec doi: 10.1101/gr.6679507 PMID:17989253 PMCID: PMC2099591 |
 |
2.
Sequencing and comparison of yeast species to identify genes and regulatory elements. (pdf)
Journal: Nature, 2003 May 15 doi: 10.1038/nature01644 PMID:12748633 |
Genes
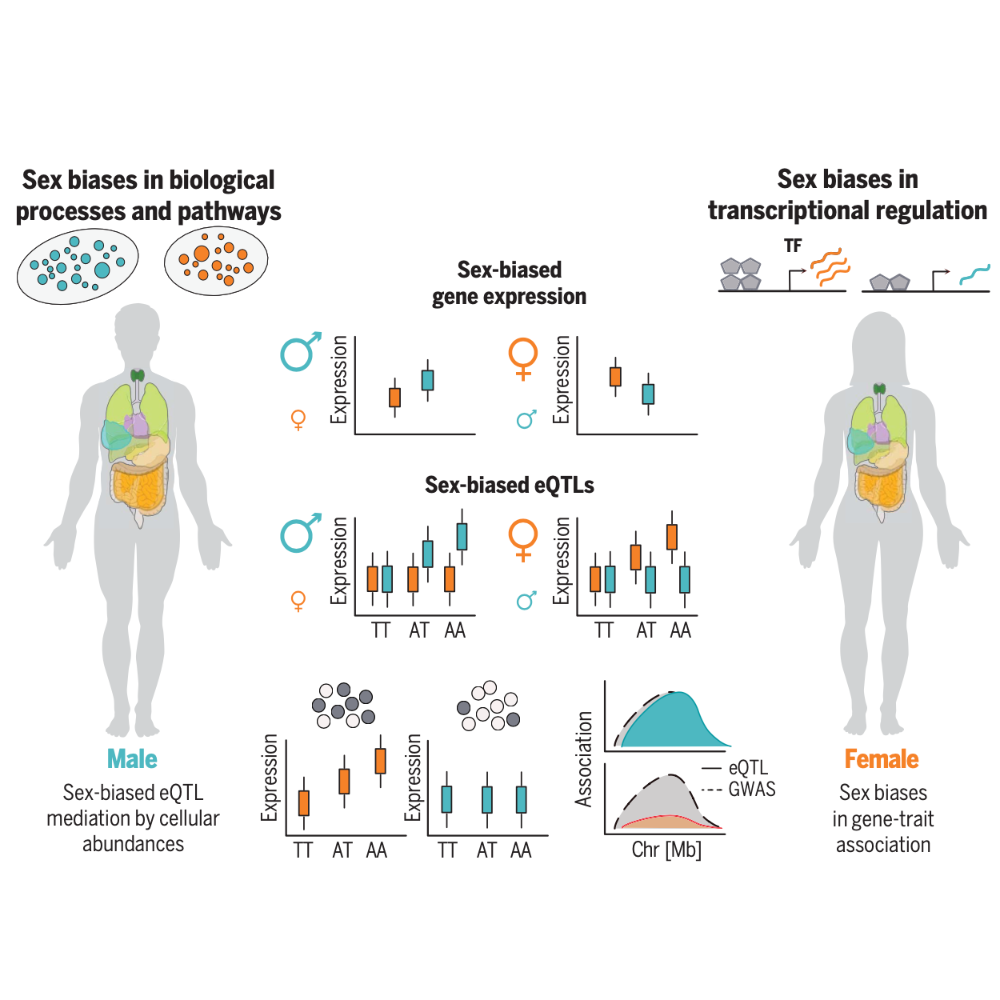 |
251.
The impact of sex on gene expression across human tissues. (pdf)
Journal: Science, 2020 Sep 11 doi: 10.1126/science.aba3066 PMID:32913072 PMCID: PMC8136152 |
 |
242.
Analysis of Genetically Regulated Gene Expression Identifies a Prefrontal PTSD Gene, SNRNP35, Specific to Military Cohorts. (pdf)
Journal: Cell Reports, 2020 Jun 2 doi: 10.1016/j.celrep.2020.107716 PMID:32492425 PMCID: PMC7359754 |
 |
157.
Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. (pdf)
Journal: Analytical Chemistry, 2016 Apr 5 doi: 10.1021/acs.analchem.6b00191 PMID:27010111 PMCID: PMC4939623 |
 |
142.
Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. (pdf)
Journal: Science, 2015 May 8 doi: 10.1126/science.1261877 PMID:25954003 PMCID: PMC4537935 |
 |
130.
FRESCo: finding regions of excess synonymous constraint in diverse viruses. (pdf)
Journal: Genome Biology, 2015 Feb 17 doi: 10.1186/s13059-015-0603-7 PMID:25853568 PMCID: PMC4376164 |
 |
118.
Evidence of efficient stop codon readthrough in four mammalian genes. (pdf)
Journal: Nucleic Acids Research, 2014 Aug doi: 10.1093/nar/gku608 PMID:25013167 PMCID: PMC4132726 |
 |
109.
Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. (pdf)
Journal: Journal Of Proteome Research, 2014 Mar 7 doi: 10.1021/pr401280w PMID:24490786 PMCID: PMC3993966 |
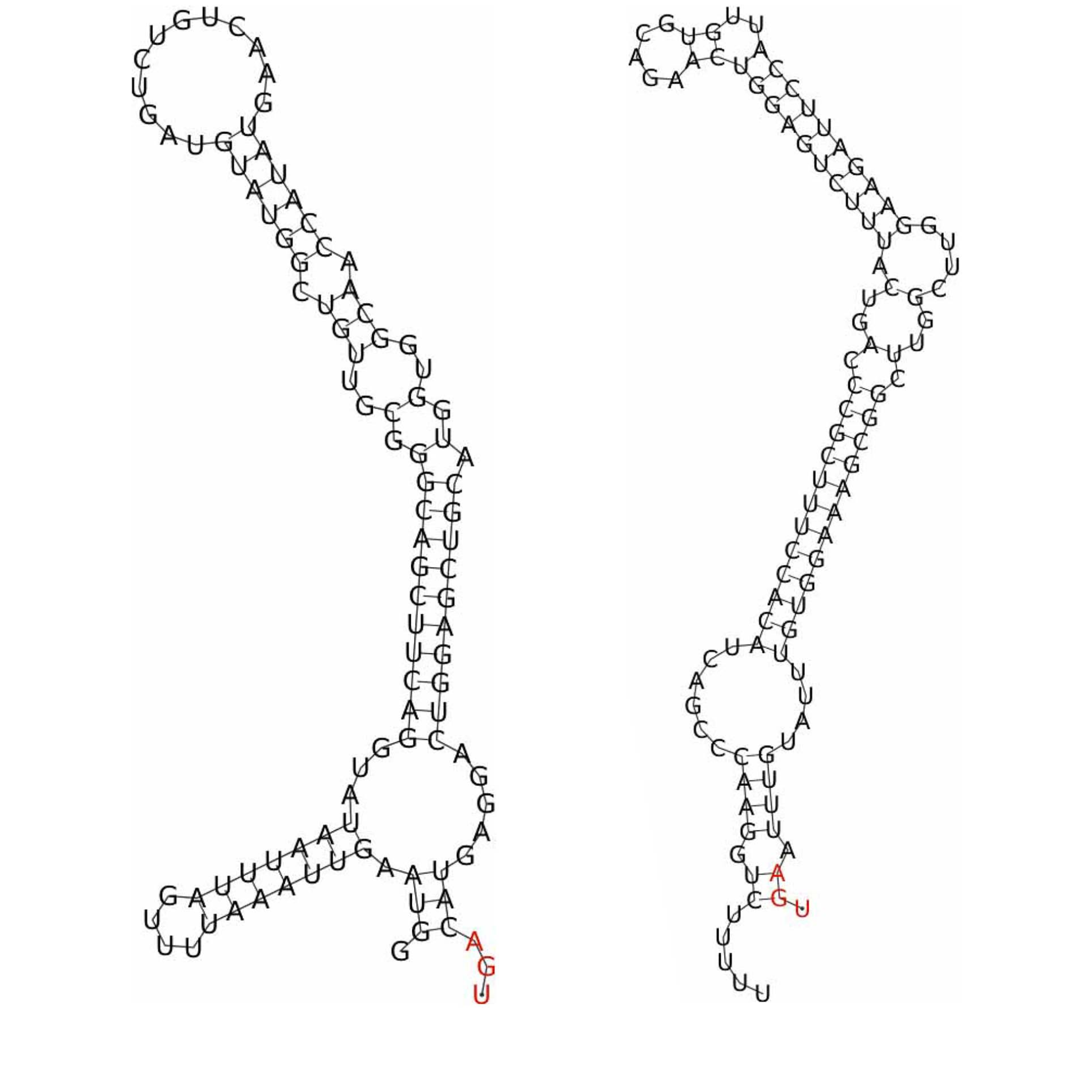 |
86.
Heterologous stop codon readthrough of metazoan readthrough candidates in yeast. (pdf)
Journal: Plos One, 2013 doi: 10.1371/journal.pone.0059450 PMID:23544069 PMCID: PMC3609751 |
 |
74.
GENCODE: the reference human genome annotation for The ENCODE Project. (pdf)
Journal: Genome Research, 2012 Sep doi: 10.1101/gr.135350.111 PMID:22955987 PMCID: PMC3431492 |
 |
60.
Evidence of abundant stop codon readthrough in Drosophila and other metazoa. (pdf)
Journal: Genome Research, 2011 Dec doi: 10.1101/gr.119974.110 PMID:21994247 PMCID: PMC3227100 |
 |
59.
Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. (pdf)
Journal: Genome Research, 2011 Nov doi: 10.1101/gr.108753.110 PMID:21994248 PMCID: PMC3205576 |
 |
41.
Optimization of parameters for coverage of low molecular weight proteins. (pdf)
Journal: Analytical And Bioanalytical Chemistry, 2010 Dec doi: 10.1007/s00216-010-4093-x PMID:20803007 PMCID: PMC2990009 |
 |
34.
The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. (pdf)
Journal: Genome Research, 2009 Jul doi: 10.1101/gr.080531.108 PMID:19498102 PMCID: PMC2704439 |
 |
26.
Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. (pdf)
Journal: Plos Computational Biology, 2008 Apr 18 doi: 10.1371/journal.pcbi.1000067 PMID:18421375 PMCID: PMC2291194 |
 |
23.
Distinguishing protein-coding and noncoding genes in the human genome. (pdf)
Journal: Proceedings Of The National Academy Of Sciences Of The United States Of America, 2007 Dec 4 doi: 10.1073/pnas.0709013104 PMID:18040051 PMCID: PMC2148306 |
 |
2.
Sequencing and comparison of yeast species to identify genes and regulatory elements. (pdf)
Journal: Nature, 2003 May 15 doi: 10.1038/nature01644 PMID:12748633 |
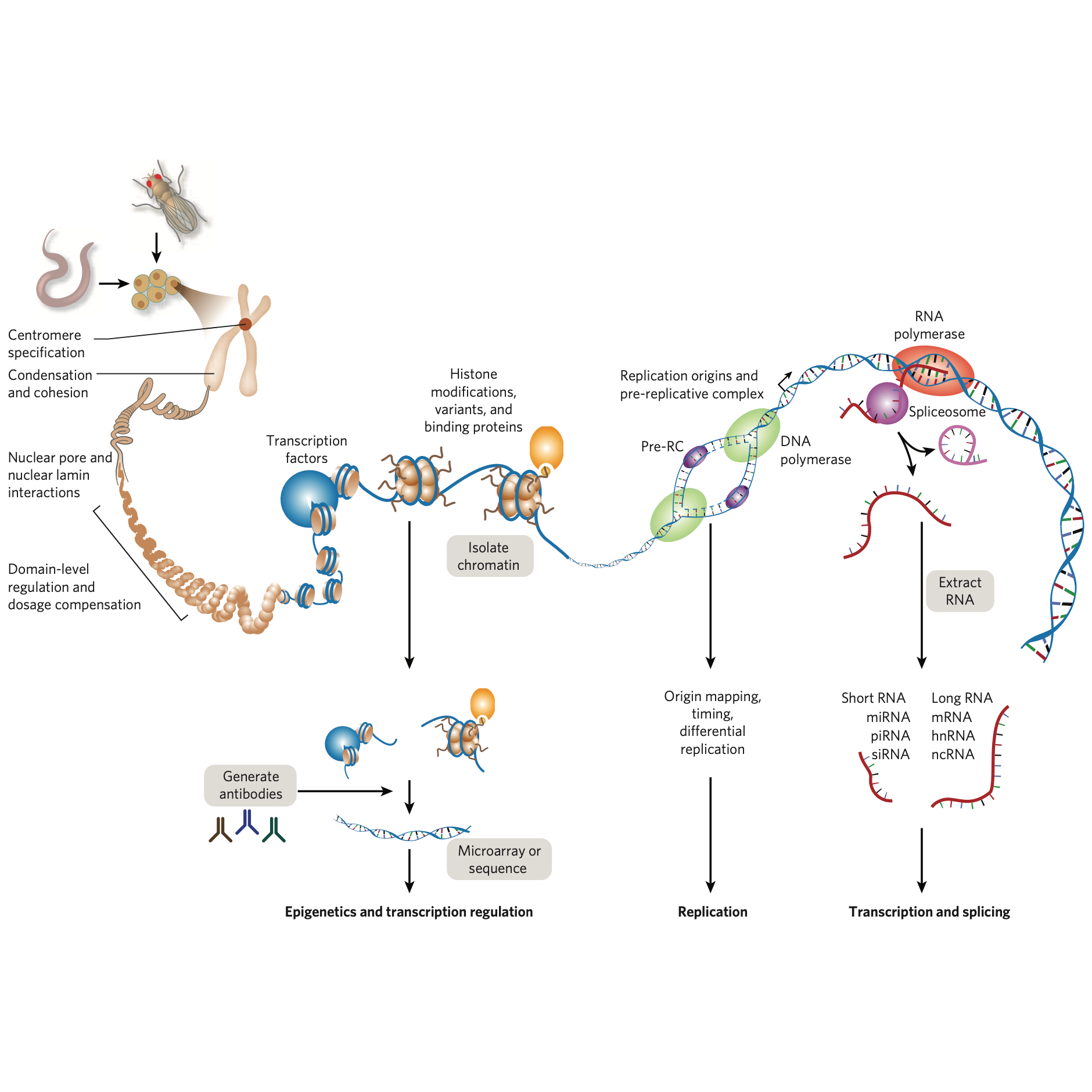
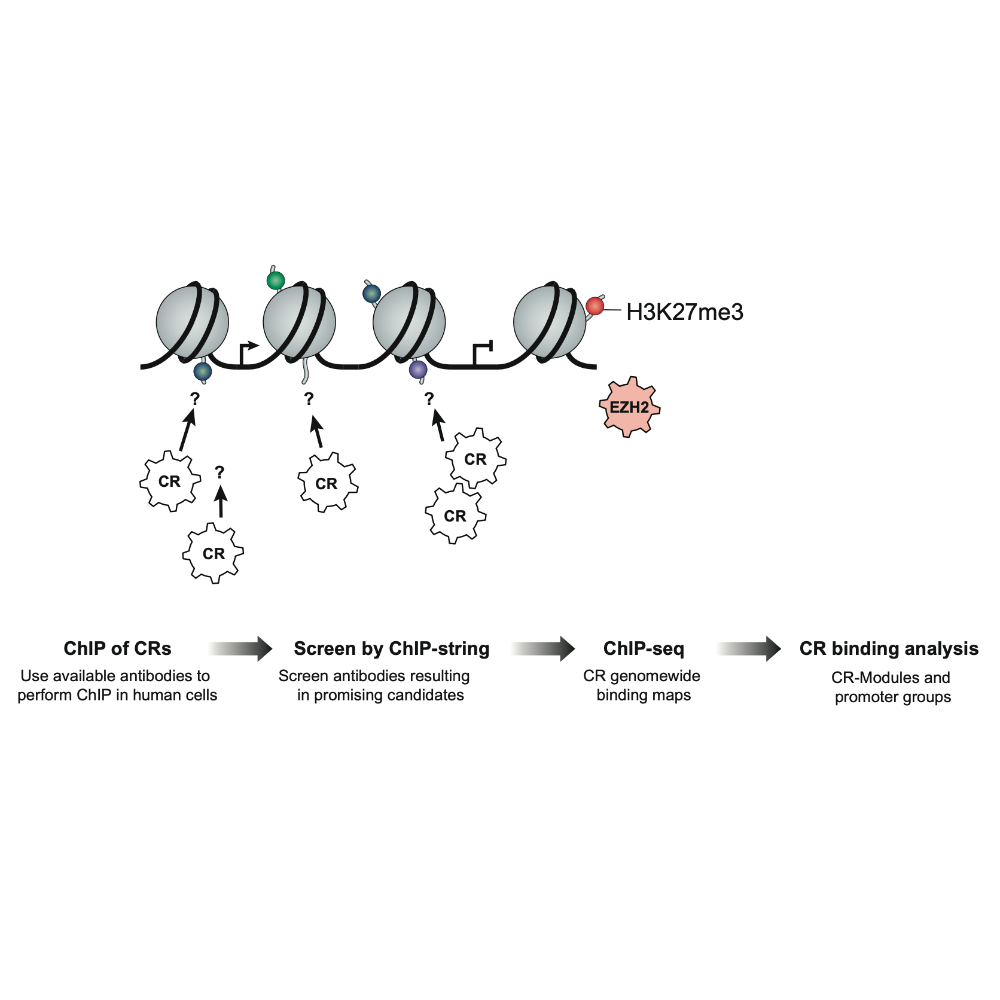
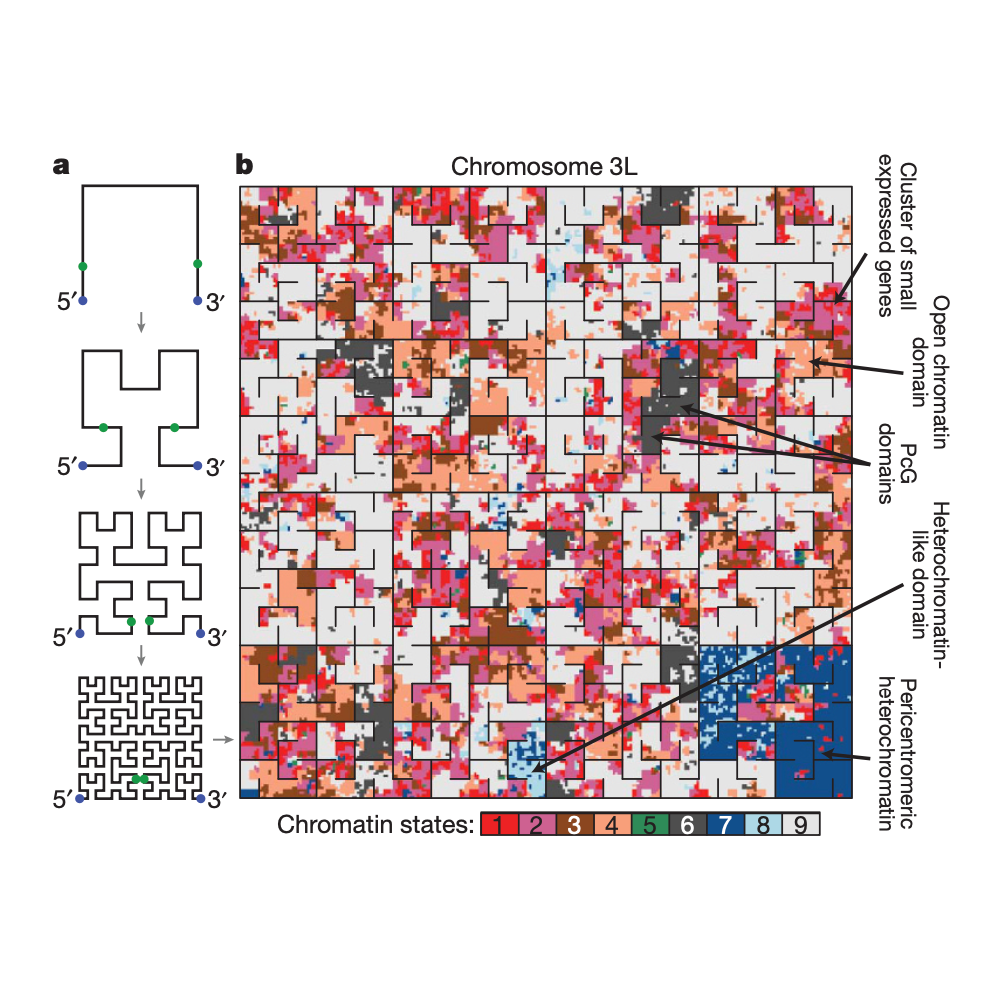
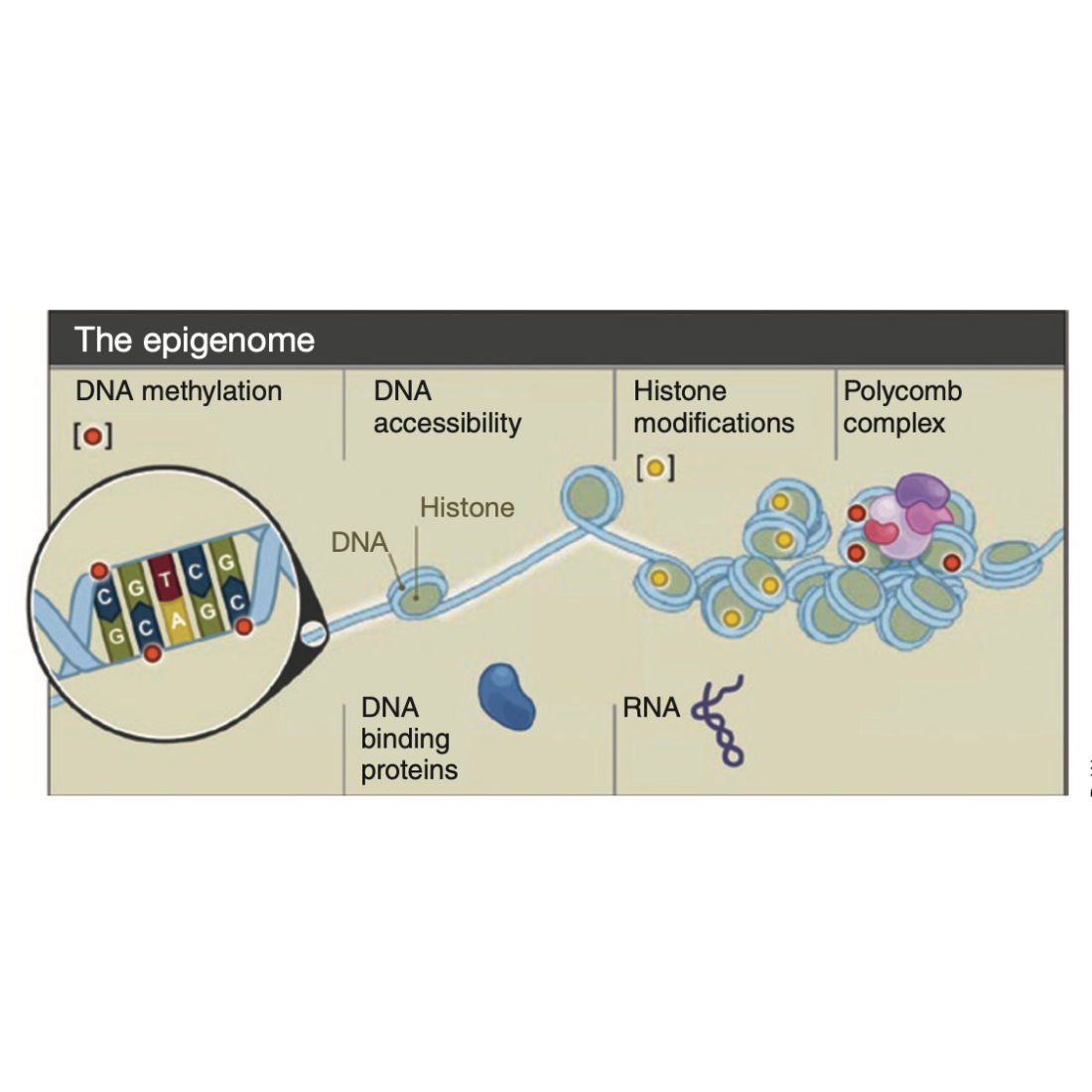
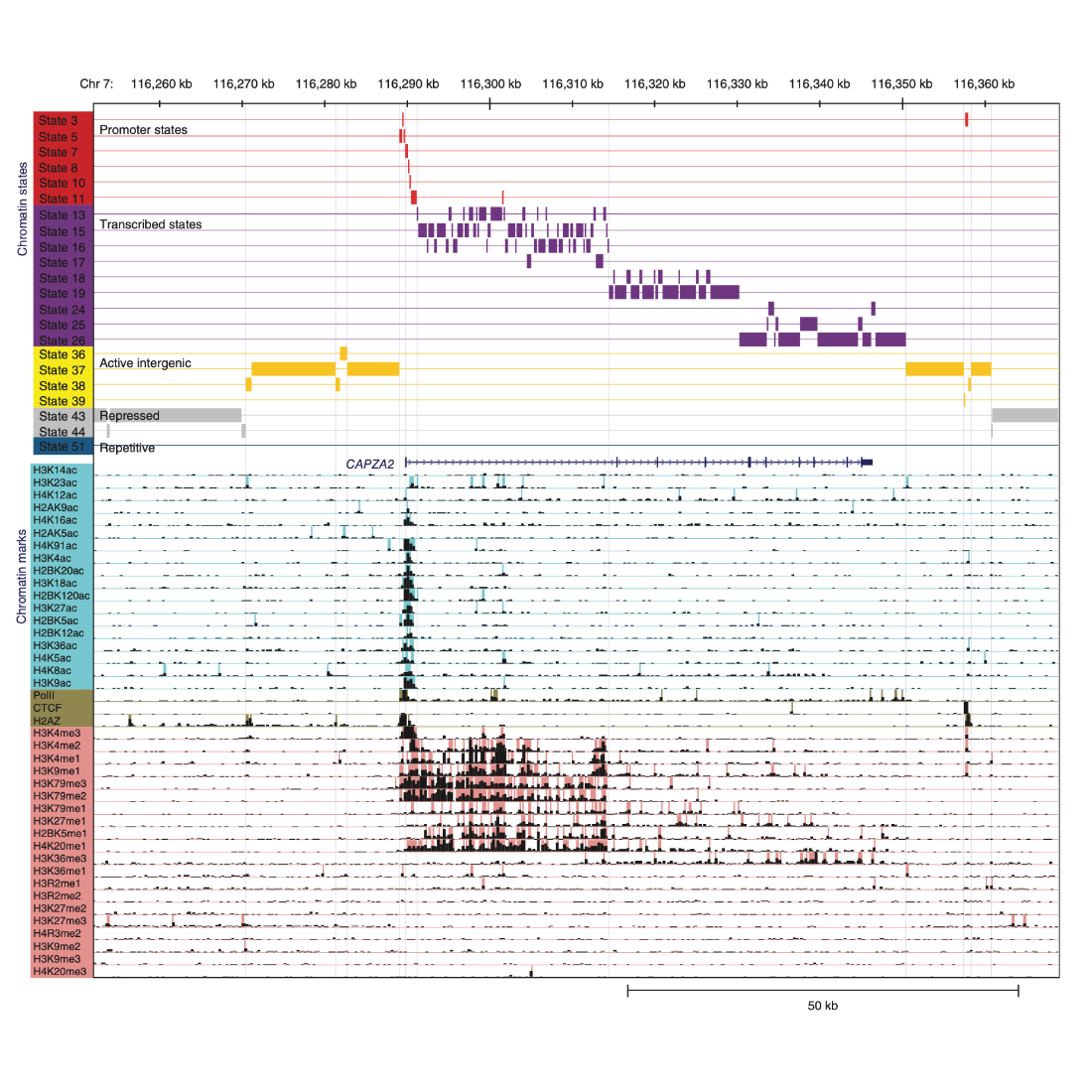
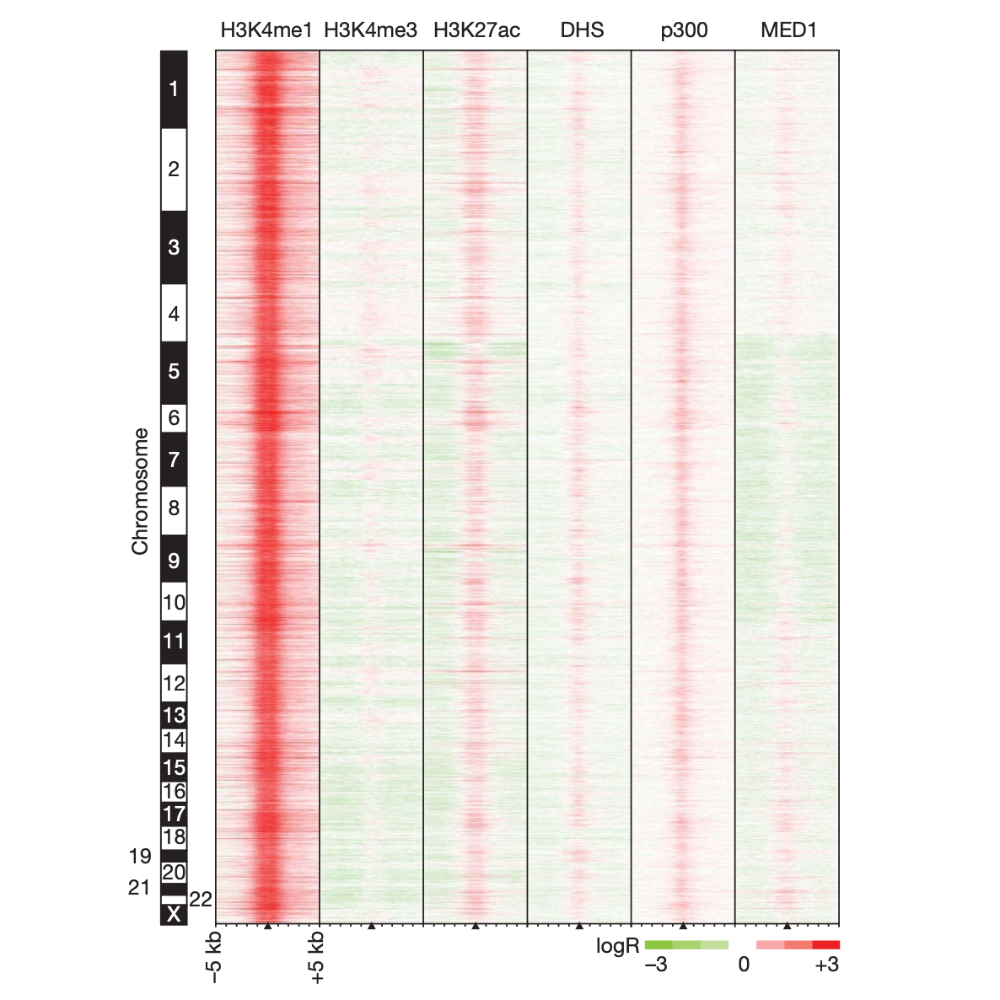
Epigenomics, Chromatin Regulation, and Developmental Programs
Beyond the primary sequence of the genome, it has recently been recognized that a wide variety of post-translational modifications in the tails of histone proteins (around which DNA is wrapped into chromatin) play key roles in genome function, cellular differentiation, and human disease. With several dozen such 'chromatin marks' already described, a wide array of chromatin regulators that can read and write them, and an astronomical number of mark combinations, discovering biologically meaningful sets of jointly acting marks and characterizing their specific function are daunting new challenges in genomics. We have developed new methods for addressing these challenges, using genome-wide maps of many histone marks to discover recurrent and spatially-coherent combinations of marks, or 'chromatin states'. We also developed data integration methods to systematically characterize chromatin state function, revealing diverse classes of enhancers, promoters, and insulators, which we used to discover new functional elements, to study chromatin dynamics across cell types and development, and to reveal motifs and regulators governing epigenetic changes in development, differentiation and disease.
 |
265.
Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. (pdf)
Journal: Molecular Cell, 2020 Dec 17 doi: 10.1016/j.molcel.2020.11.041 PMID:33290746 PMCID: PMC7691212 |
 |
205.
A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS). (pdf)
Journal: Nature Communications, 2019 Jun 28 doi: 10.1038/s41467-019-10912-8 PMID:31253799 PMCID: PMC6599391 |
 |
198.
An AR-ERG transcriptional signature defined by long-range chromatin interactomes in prostate cancer cells. (pdf)
Journal: Genome Research, 2019 Feb doi: 10.1101/gr.230243.117 PMID:30606742 PMCID: PMC6360806 |
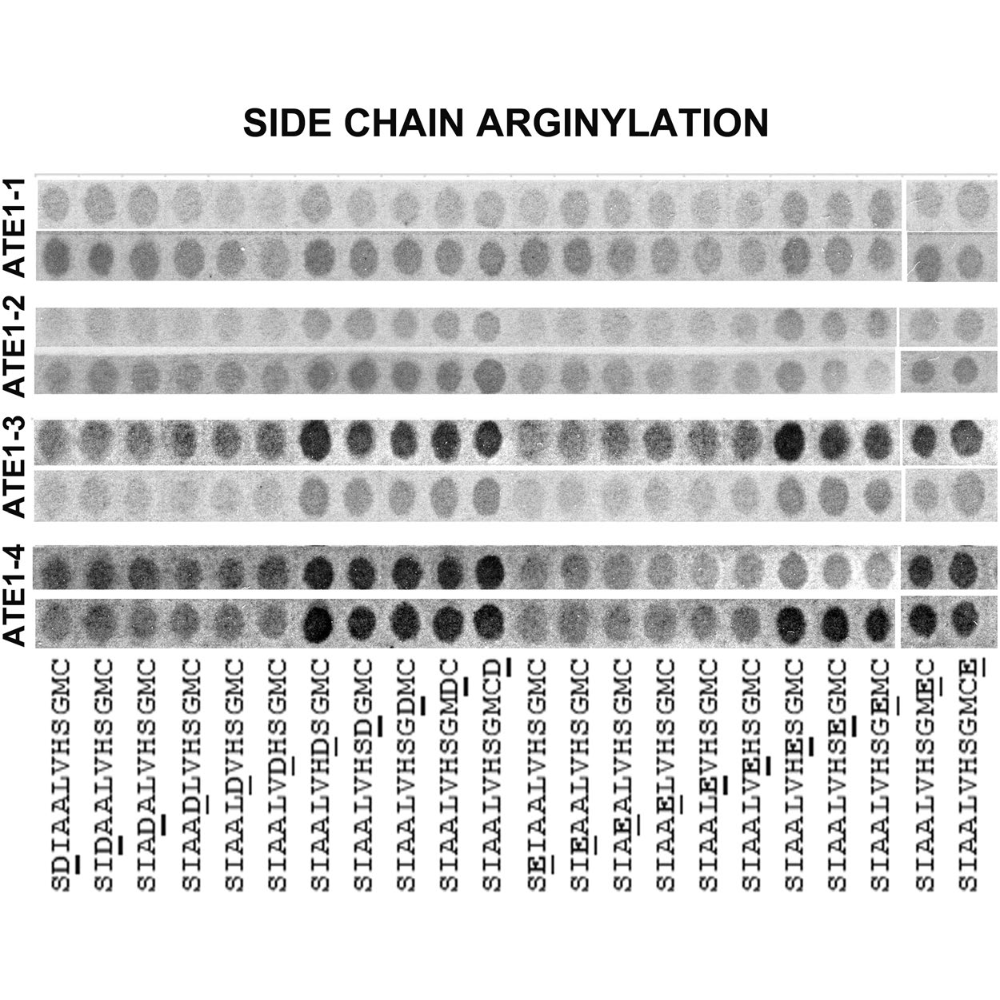 |
192.
Target site specificity and in vivo complexity of the mammalian arginylome. (pdf)
Journal: Scientific Reports, 2018 Nov 1 doi: 10.1038/s41598-018-34639-6 PMID:30385798 PMCID: PMC6212499 |
 |
191.
Allele-specific epigenome maps reveal sequence-dependent stochastic switching at regulatory loci. (pdf)
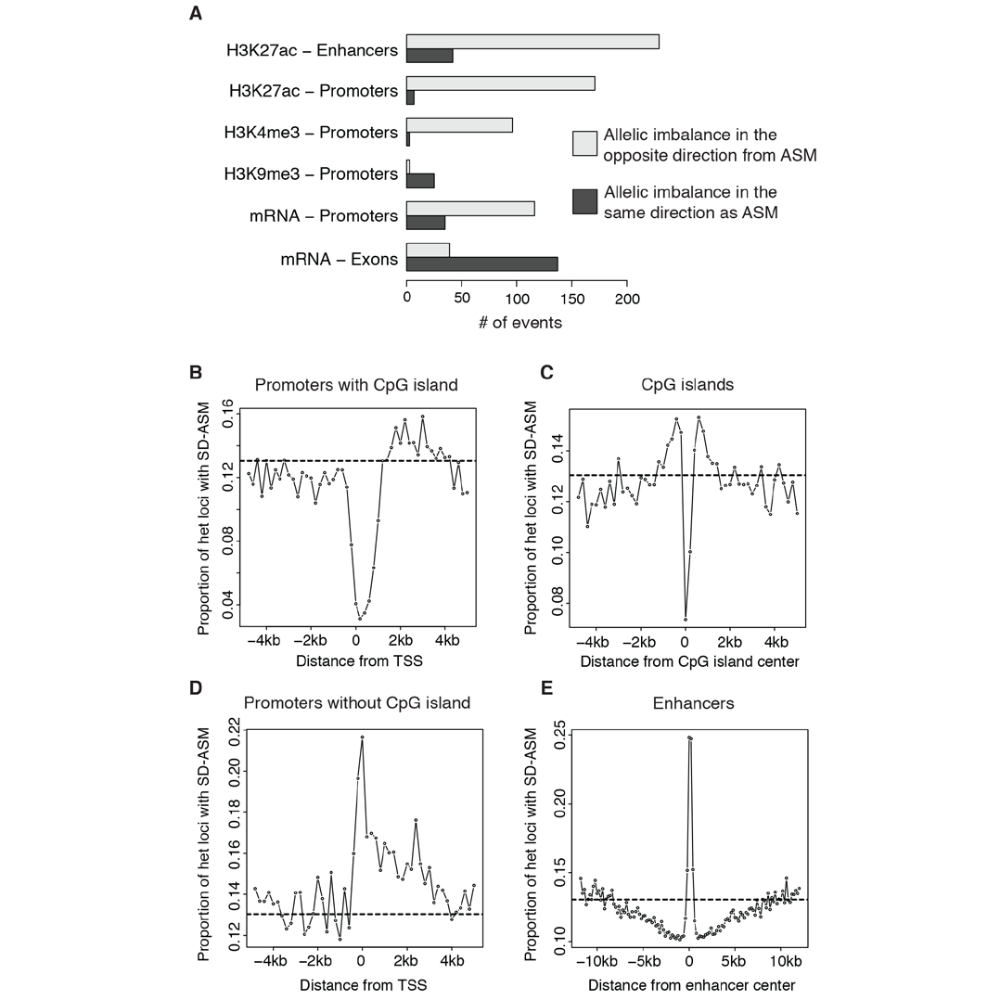
Journal: Science, 2018 Sep 28 doi: 10.1126/science.aar3146 PMID:30139913 PMCID: PMC6198826 |
 |
184.
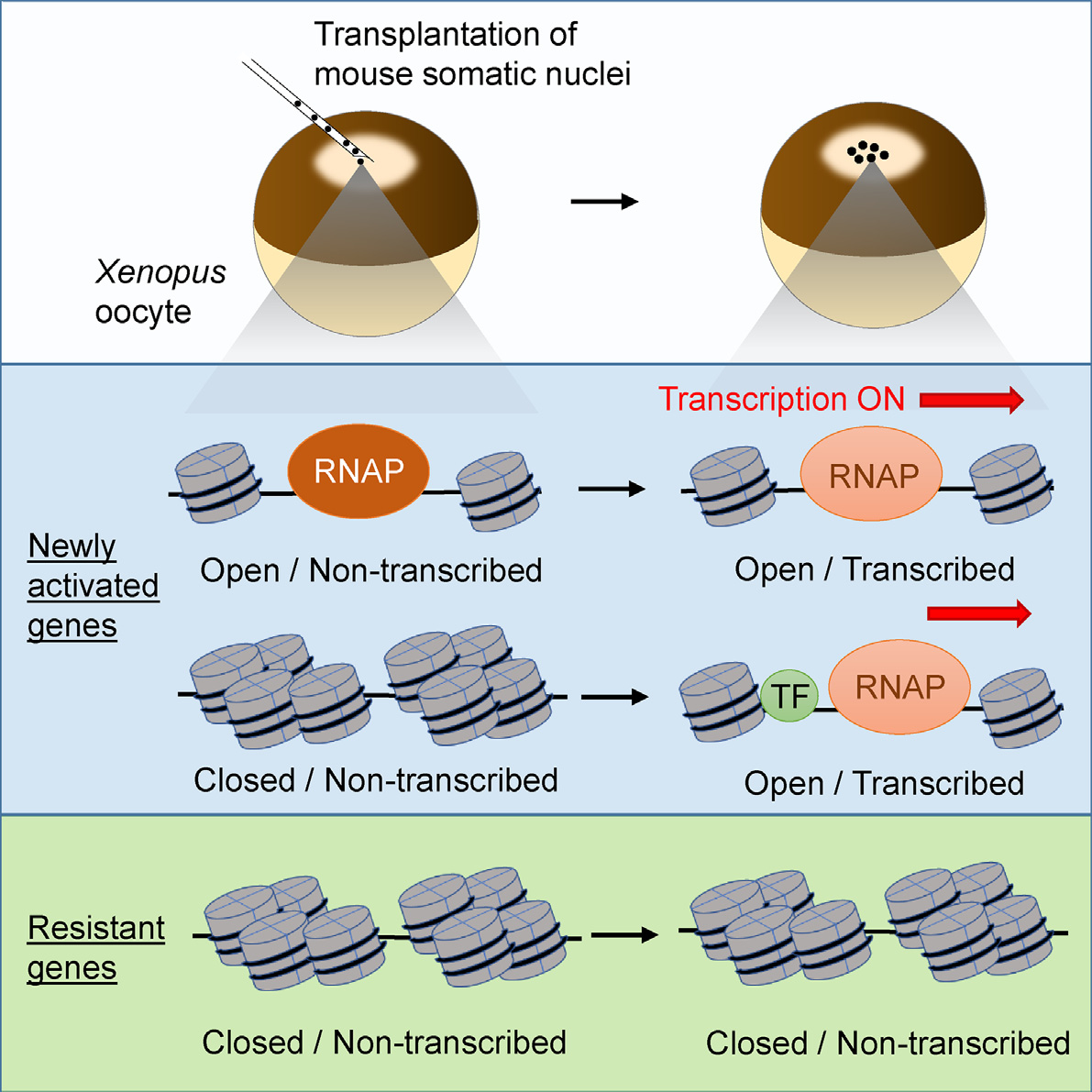
Chromatin Accessibility Impacts Transcriptional Reprogramming in Oocytes. (pdf)
Journal: Cell Reports, 2018 Jul 10 doi: 10.1016/j.celrep.2018.06.030 PMID:29996092 PMCID: PMC6057489 |
 |
177.
Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. (pdf)
Journal: Nature Genetics, 2017 Dec doi: 10.1038/ng.3969 PMID:29019975 PMCID: PMC6636856 |
 |
174.
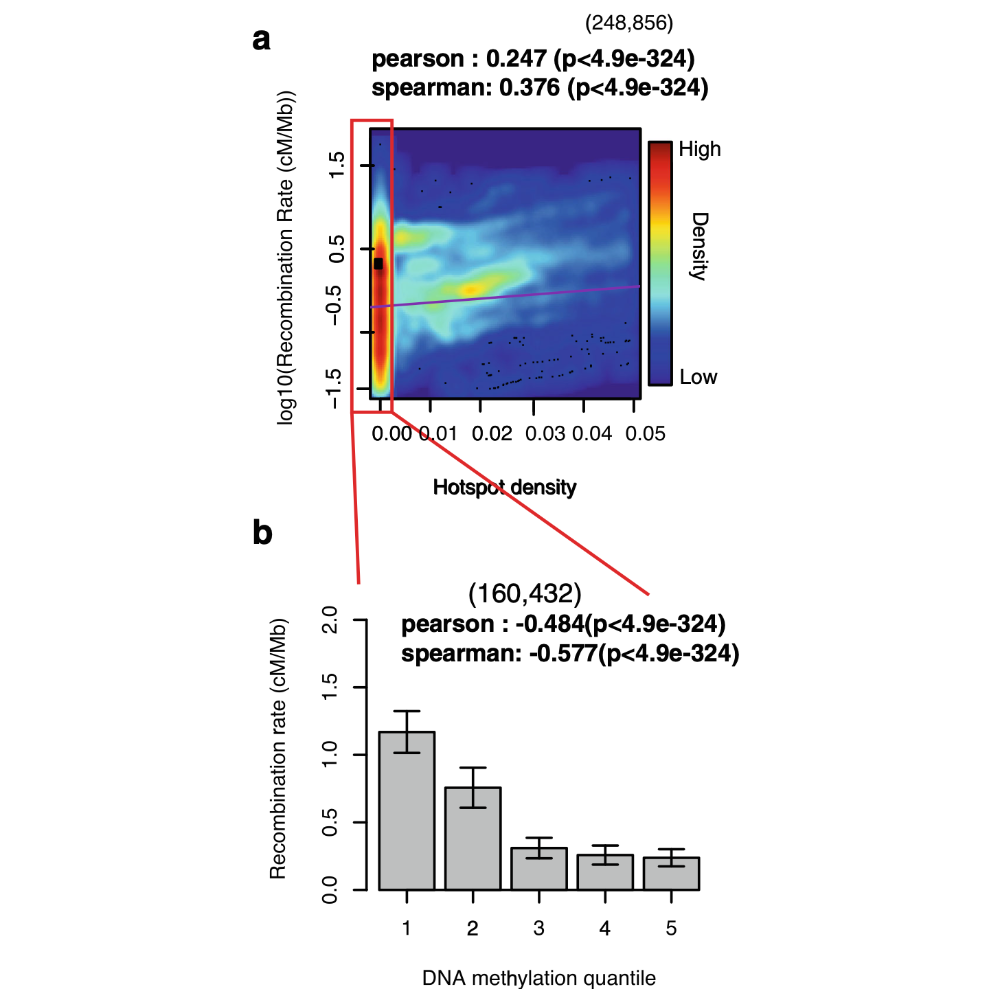
Evidence of reduced recombination rate in human regulatory domains. (pdf)
Journal: Genome Biology, 2017 Oct 20 doi: 10.1186/s13059-017-1308-x PMID:29058599 PMCID: PMC5651596 |
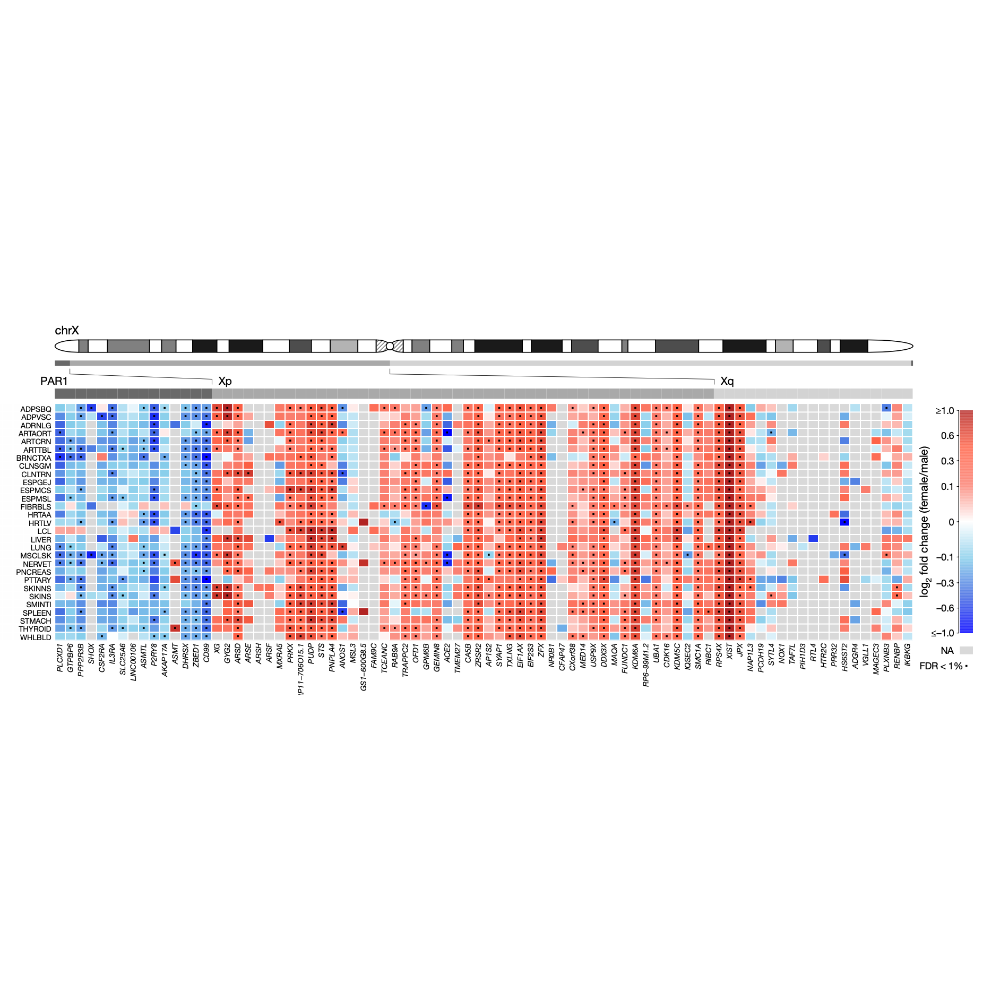 |
170.
Landscape of X chromosome inactivation across human tissues. (pdf)
Journal: Nature, 2017 Oct 11 doi: 10.1038/nature24265 PMID:29022598 PMCID: PMC5685192 |
 |
168.
Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. (pdf)
Journal: Nature Genetics, 2017 Jun doi: 10.1038/ng.3841 PMID:28436984 PMCID: PMC5841952 |
 |
167.
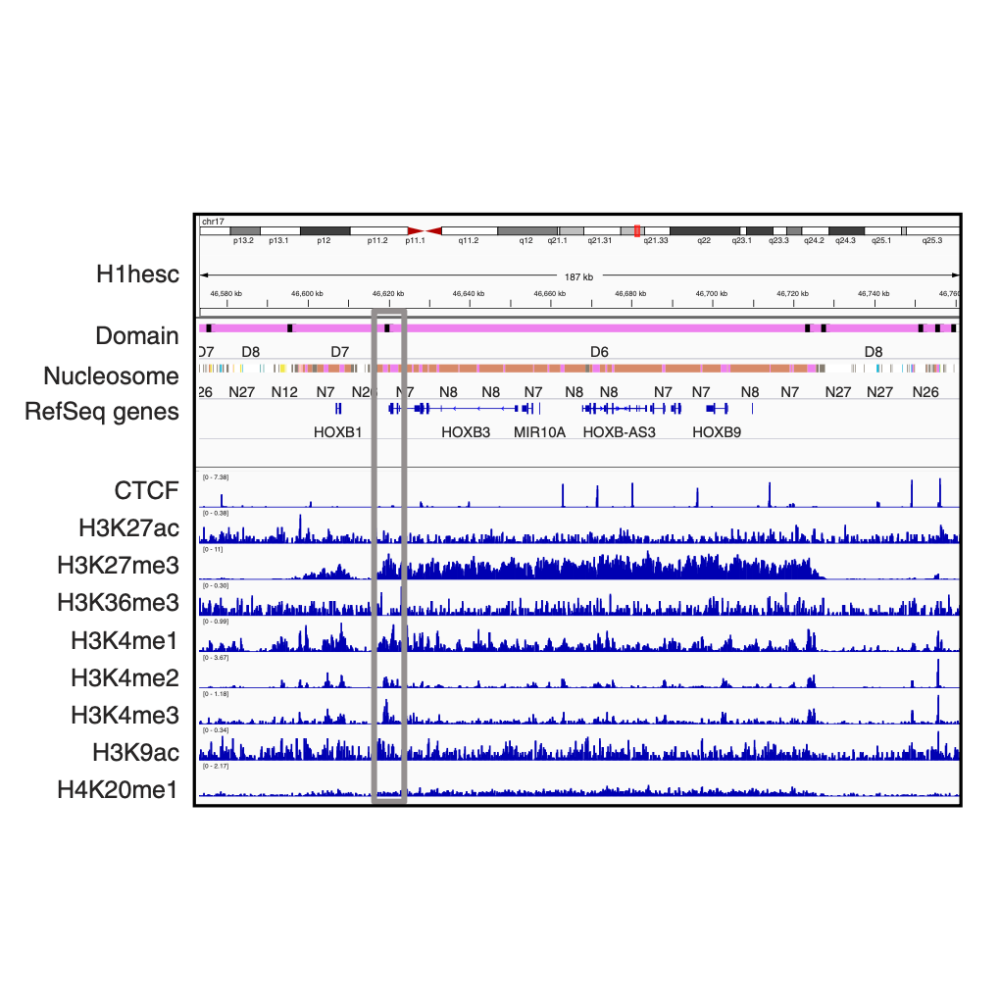
Multi-scale chromatin state annotation using a hierarchical hidden Markov model. (pdf)
Journal: Nature Communications, 2017 Apr 7 doi: 10.1038/ncomms15011 PMID:28387224 PMCID: PMC5385569 |
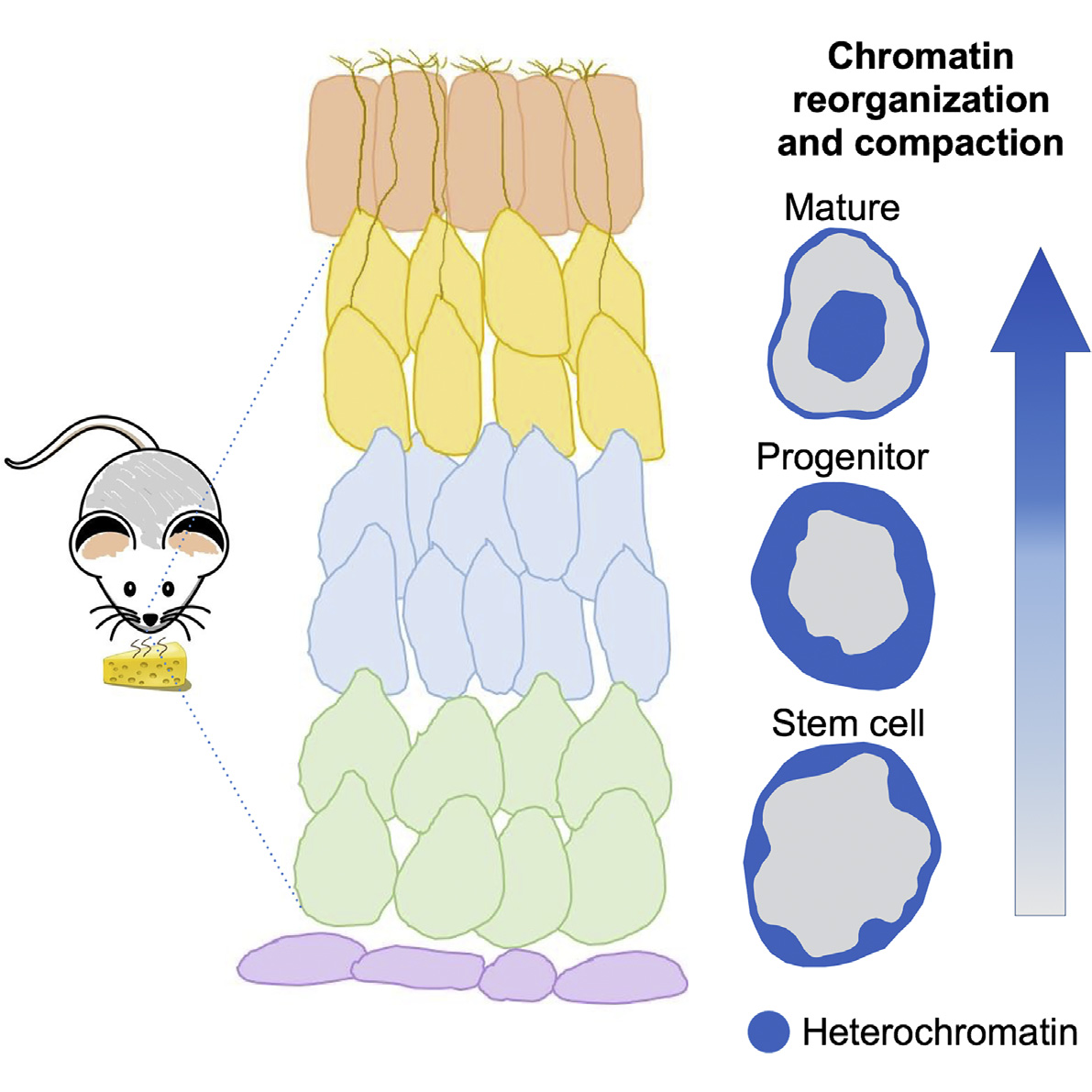 |
164.
Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. (pdf)
Journal: Cell Reports, 2016 Nov 15 doi: 10.1016/j.celrep.2016.10.060 PMID:27851973 PMCID: PMC5135017 |
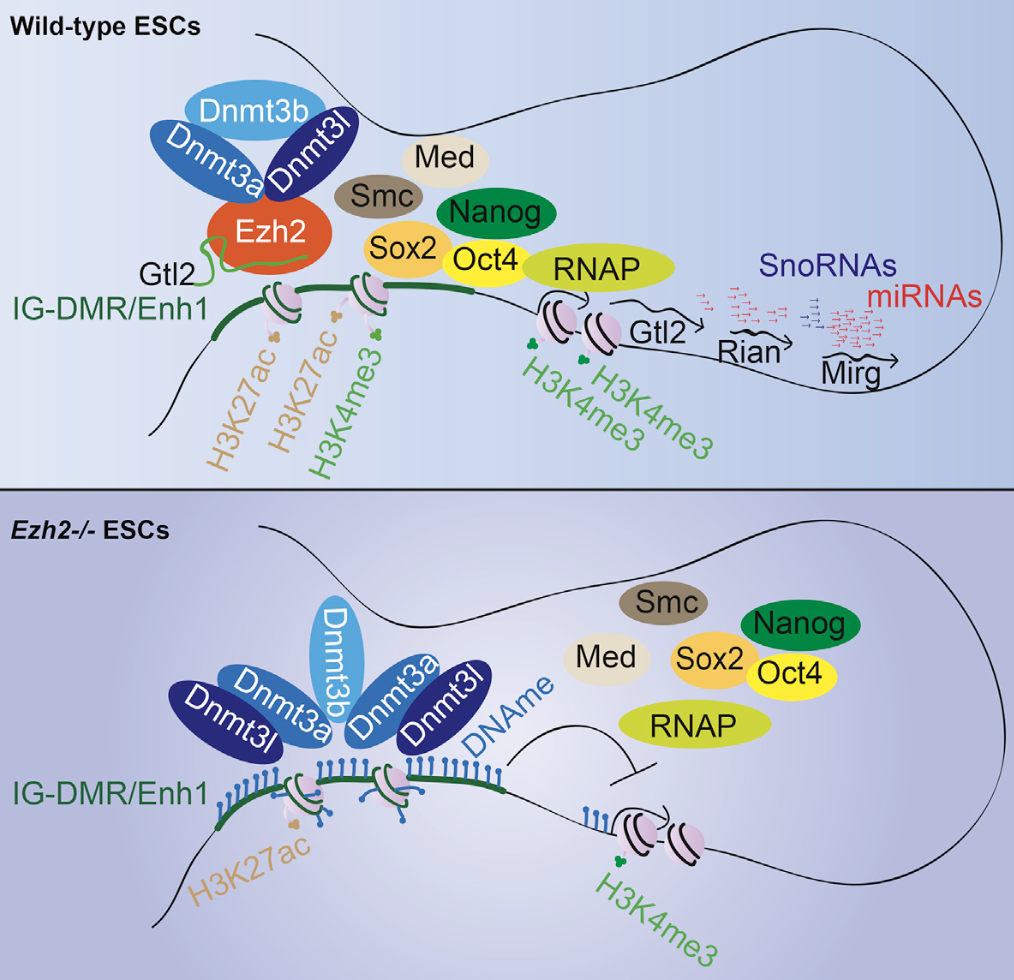 |
148.
PRC2 Is Required to Maintain Expression of the Maternal Gtl2-Rian-Mirg Locus by Preventing De Novo DNA Methylation in Mouse Embryonic Stem Cells. (pdf)
Journal: Cell Reports, 2015 Sep 1 doi: 10.1016/j.celrep.2015.07.053 PMID:26299972 PMCID: PMC5384103 |
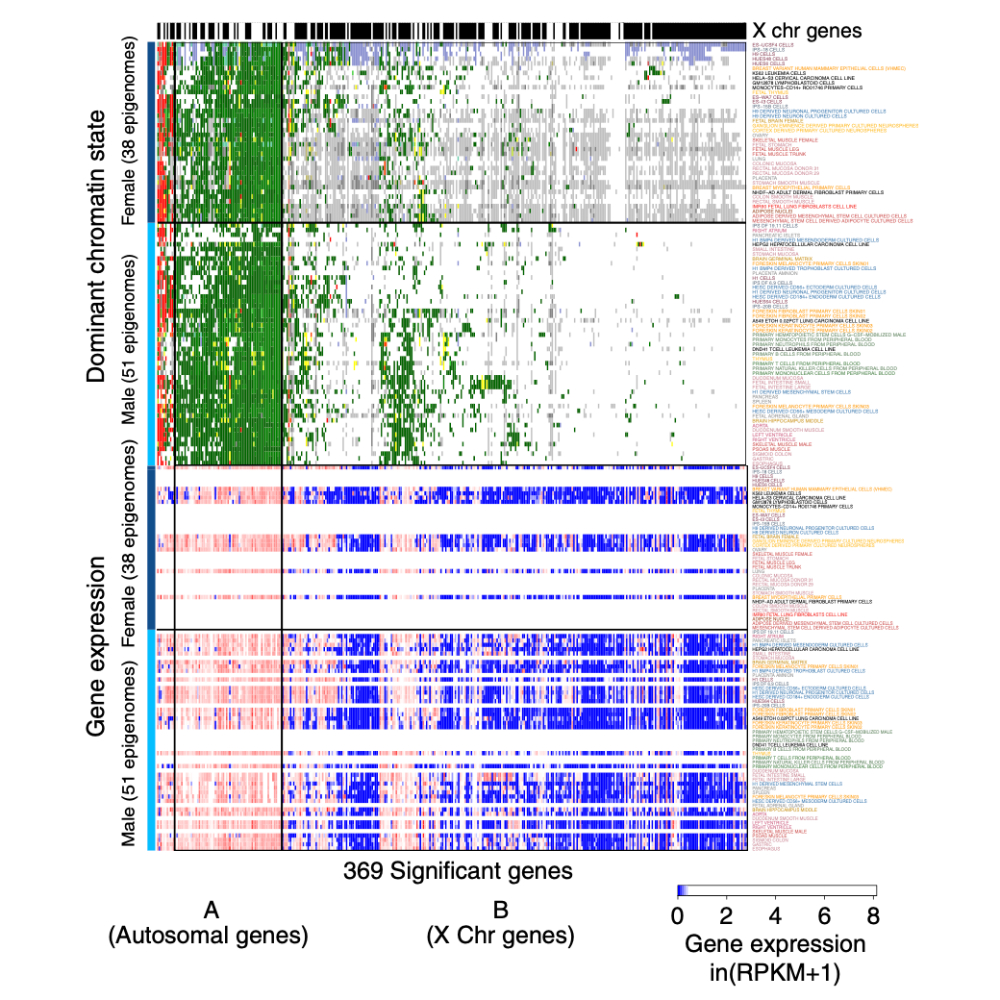 |
147.
Systematic chromatin state comparison of epigenomes associated with diverse properties including sex and tissue type. (pdf)
Journal: Nature Communications, 2015 Aug 18 doi: 10.1038/ncomms8973 PMID:26282110 PMCID: PMC4557131 |
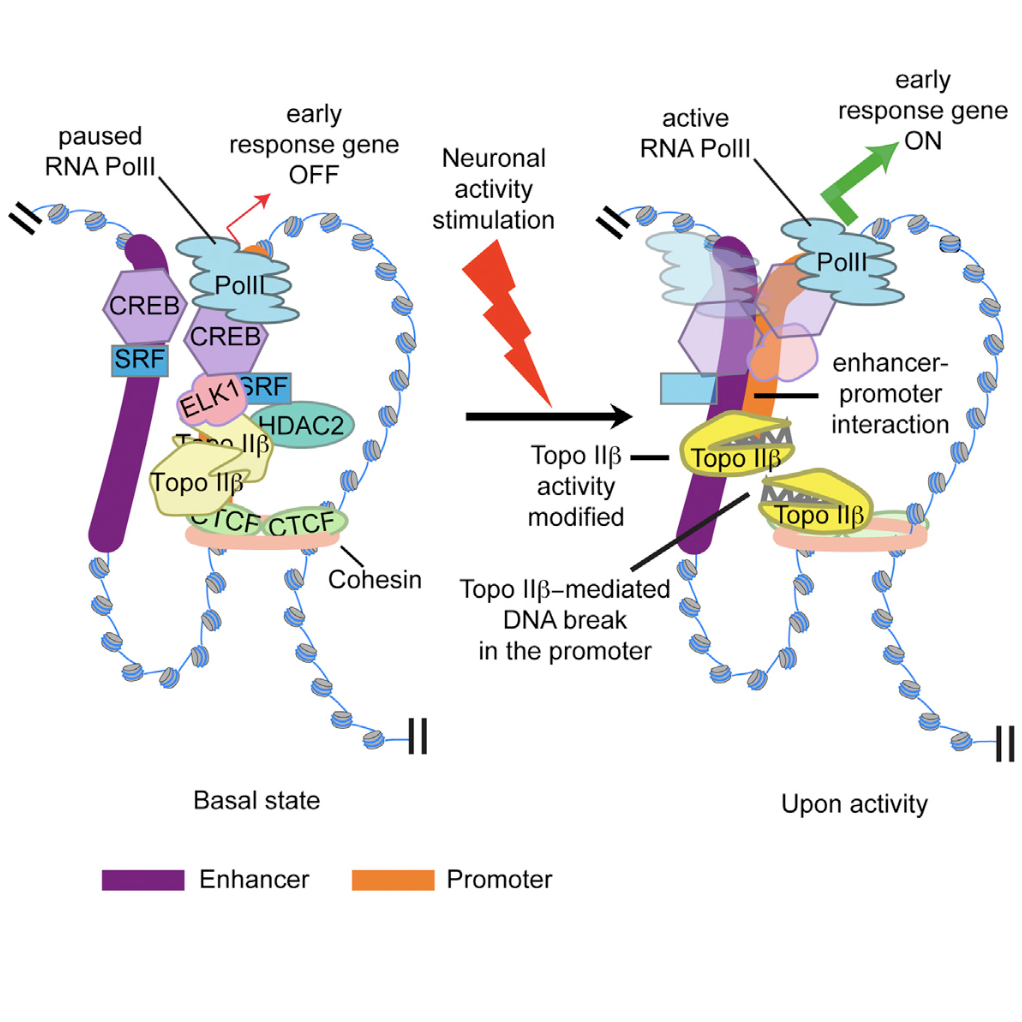 |
145.
Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. (pdf)
Journal: Cell, 2015 Jun 18 doi: 10.1016/j.cell.2015.05.032 PMID:26052046 PMCID: PMC4886855 |
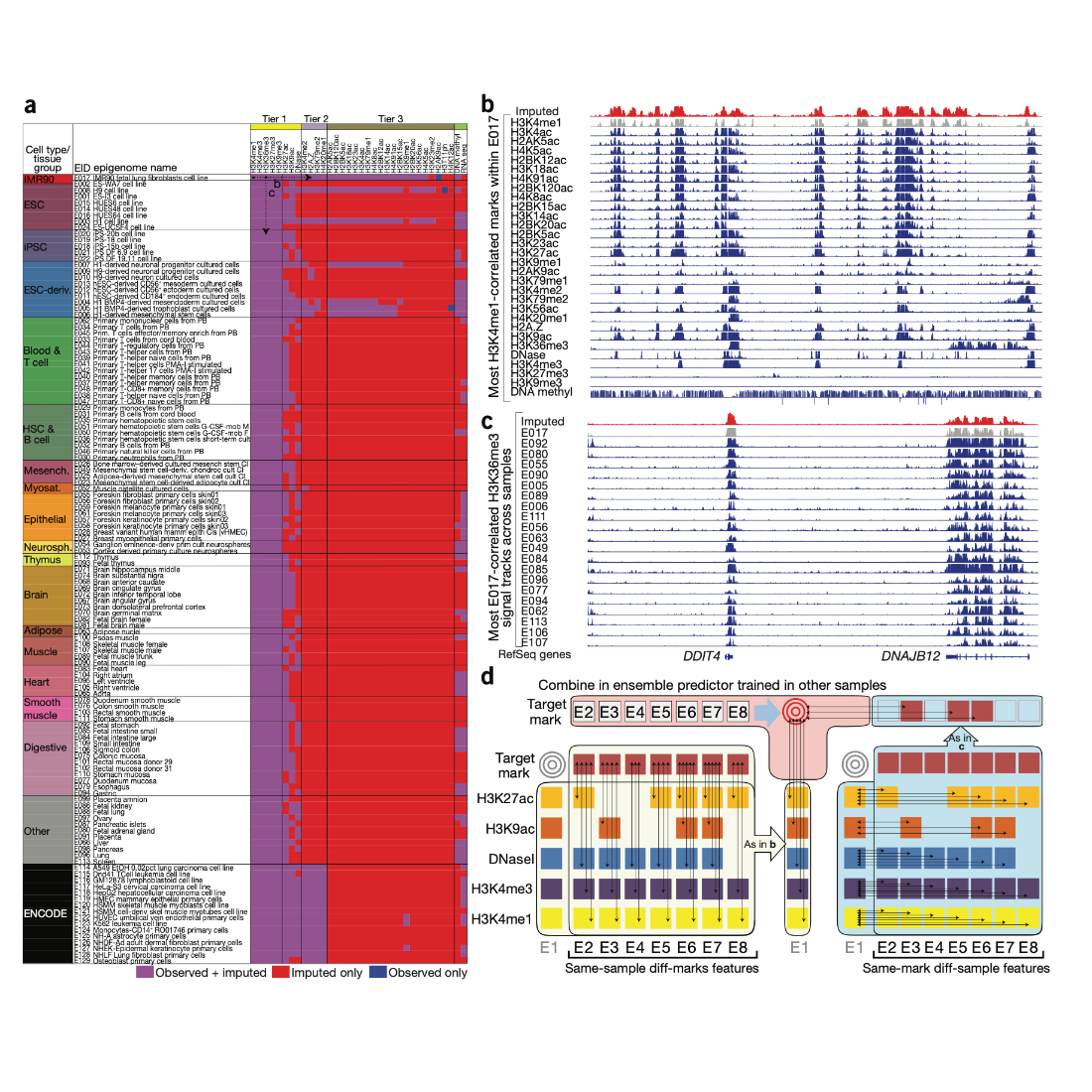 |
138.
Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. (pdf)
Journal: Nature Biotechnology, 2015 Apr doi: 10.1038/nbt.3157 PMID:25690853 PMCID: PMC4512306 |
 |
134.
BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. (pdf)
Journal: Molecular Cell, 2015 Feb 19 doi: 10.1016/j.molcel.2015.01.011 PMID:25699710 PMCID: PMC4351672 |
 |
132.
Integrative analysis of 111 reference human epigenomes. (pdf)
Journal: Nature, 2015 Feb 19 doi: 10.1038/nature14248 PMID:25693563 PMCID: PMC4530010 |
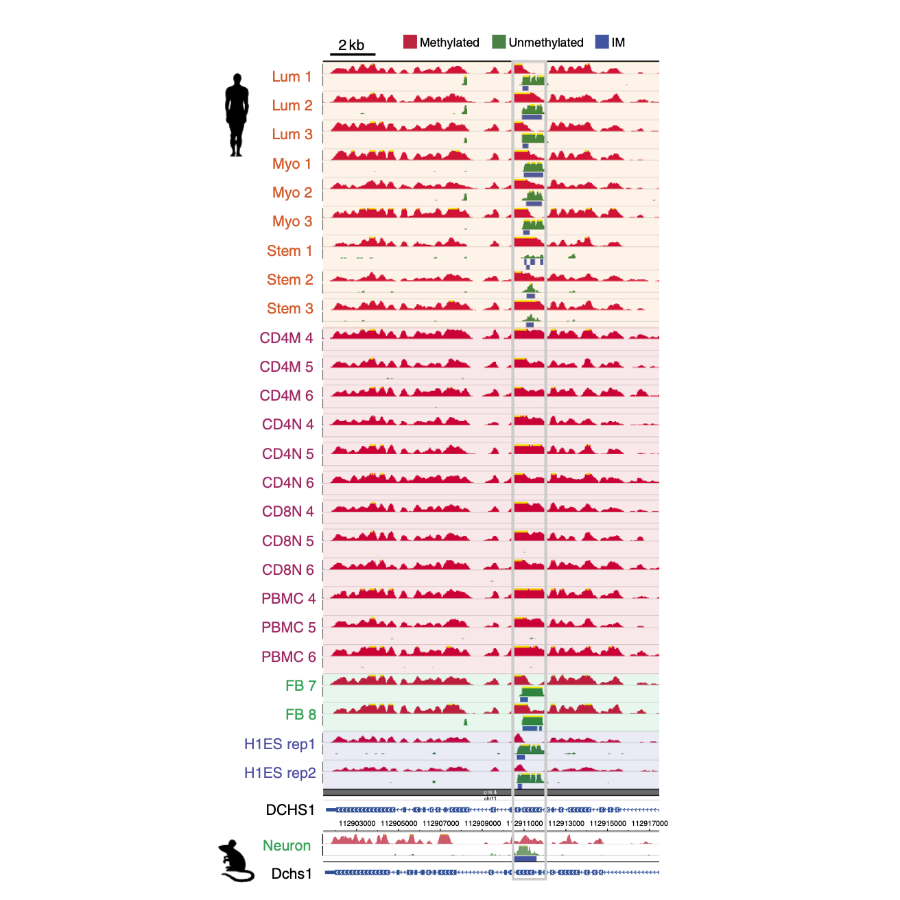 |
131.
Intermediate DNA methylation is a conserved signature of genome regulation. (pdf)
Journal: Nature Communications, 2015 Feb 18 doi: 10.1038/ncomms7363 PMID:25691127 PMCID: PMC4333717 |
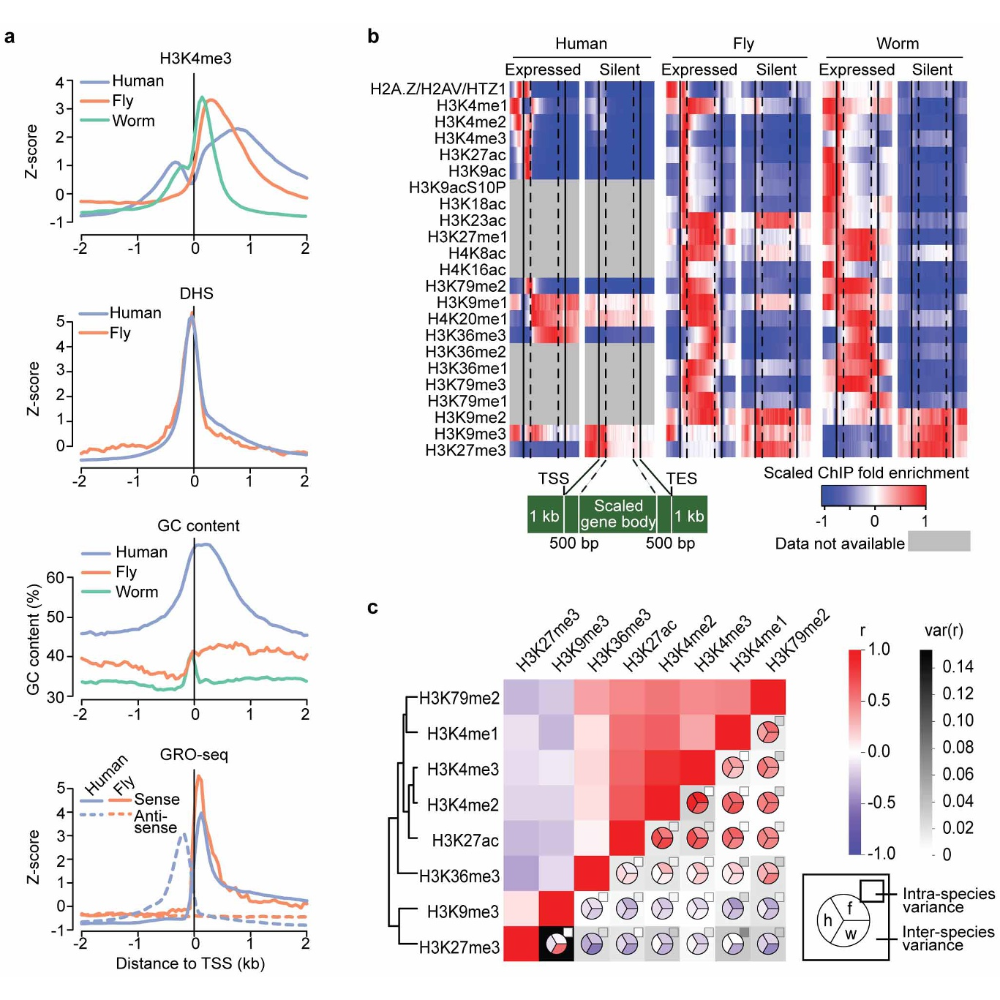 |
122.
Comparative analysis of metazoan chromatin organization. (pdf)
Journal: Nature, 2014 Aug 28 doi: 10.1038/nature13415 PMID:25164756 PMCID: PMC4227084 |
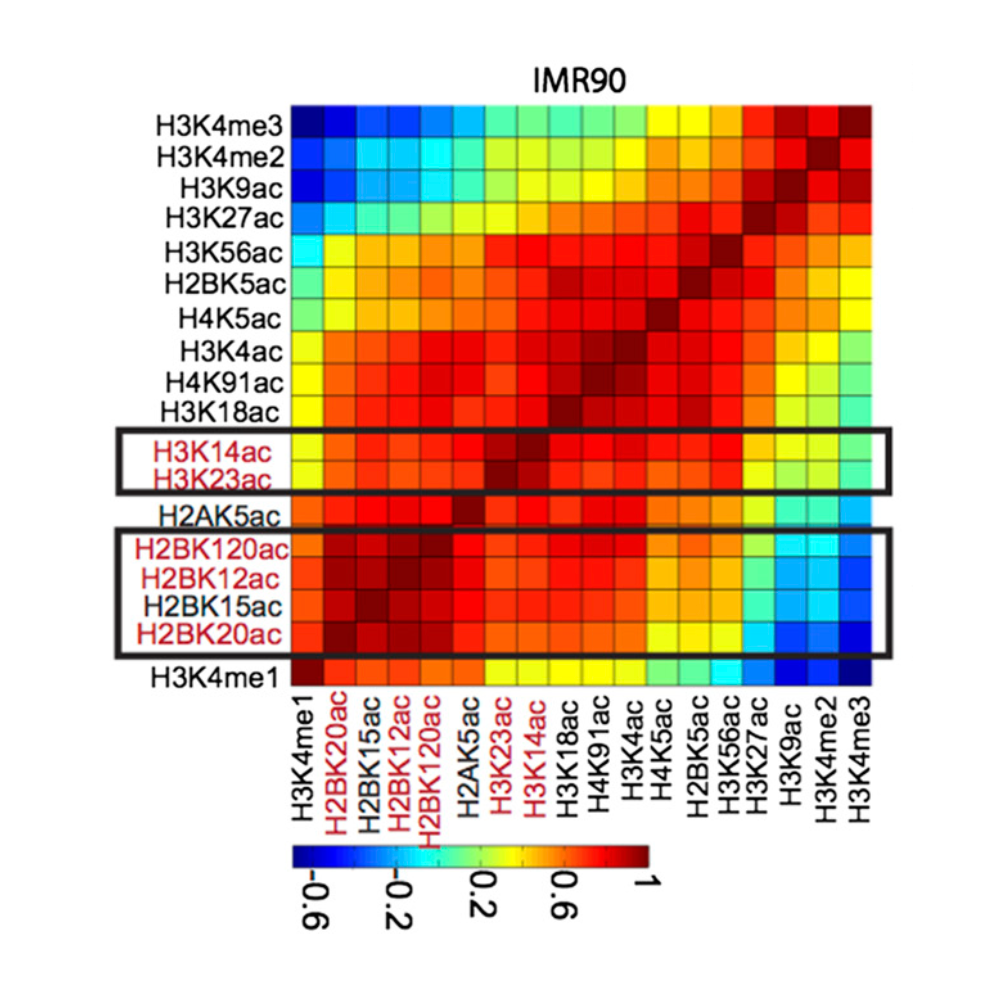 |
119.
Distinct and predictive histone lysine acetylation patterns at promoters, enhancers, and gene bodies. (pdf)
Journal: G3, 2014 Aug 12 doi: 10.1534/g3.114.013565 PMID:25122670 PMCID: PMC4232531 |
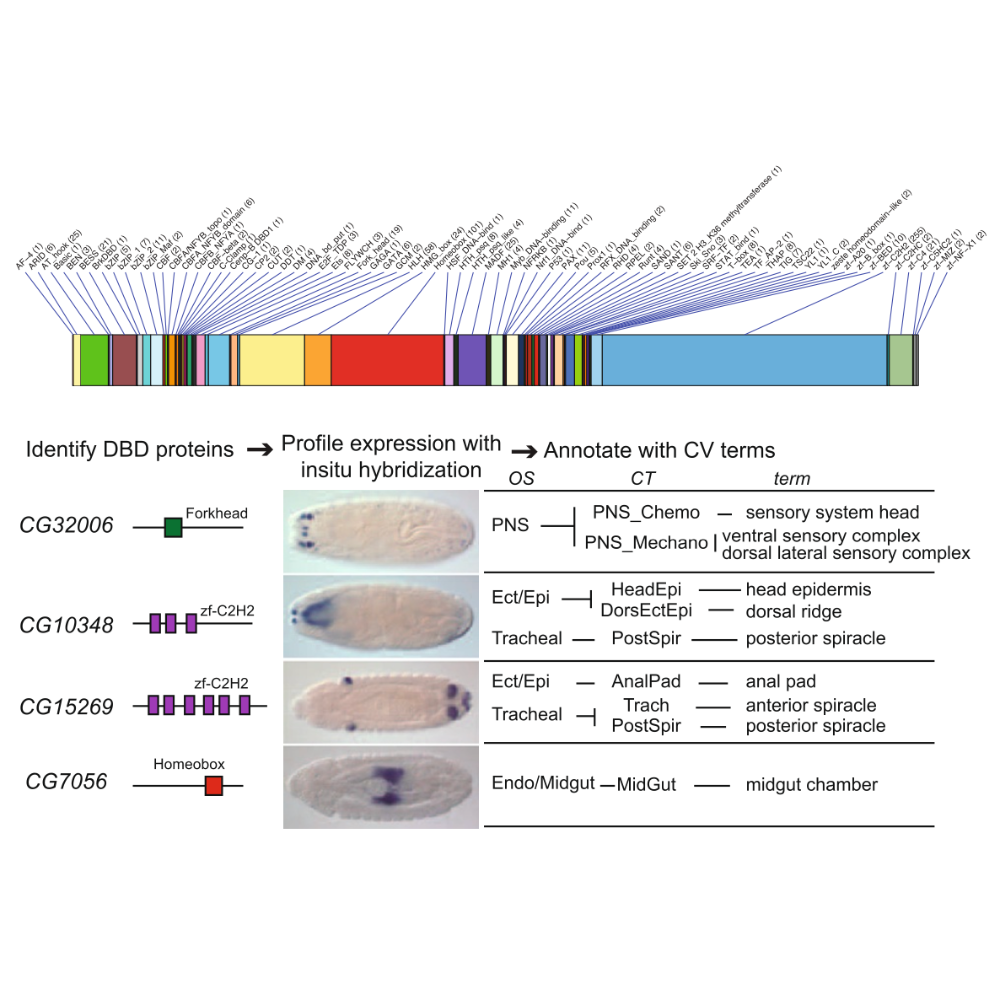 |
103.
Spatial expression of transcription factors in Drosophila embryonic organ development. (pdf)
Journal: Genome Biology, 2013 Dec 20 doi: 10.1186/gb-2013-14-12-r140 PMID:24359758 PMCID: PMC4053779 |
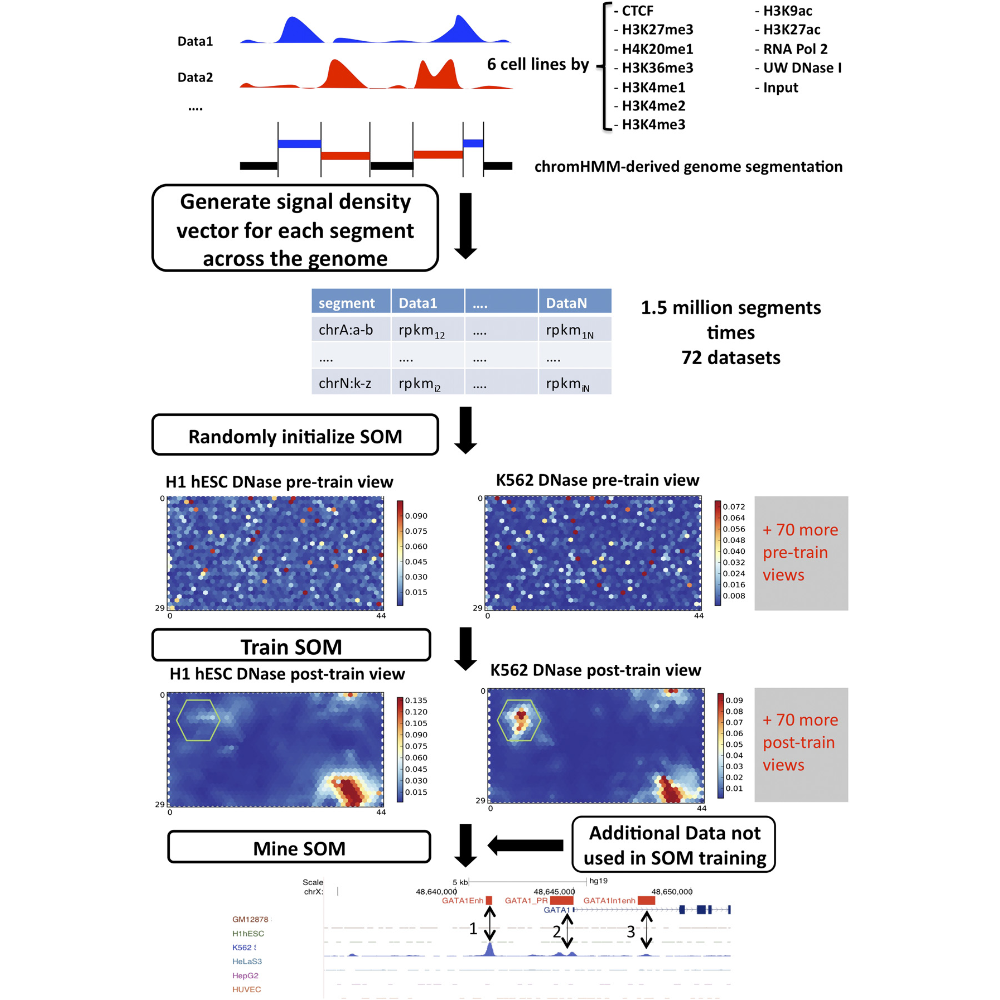 |
102.
Integrating and mining the chromatin landscape of cell-type specificity using self-organizing maps. (pdf)
Journal: Genome Research, 2013 Dec doi: 10.1101/gr.158261.113 PMID:24170599 PMCID: PMC3847782 |
 |
101.
Extensive variation in chromatin states across humans. (pdf)
Journal: Science, 2013 Nov 8 doi: 10.1126/science.1242510 PMID:24136358 PMCID: PMC4075767 |
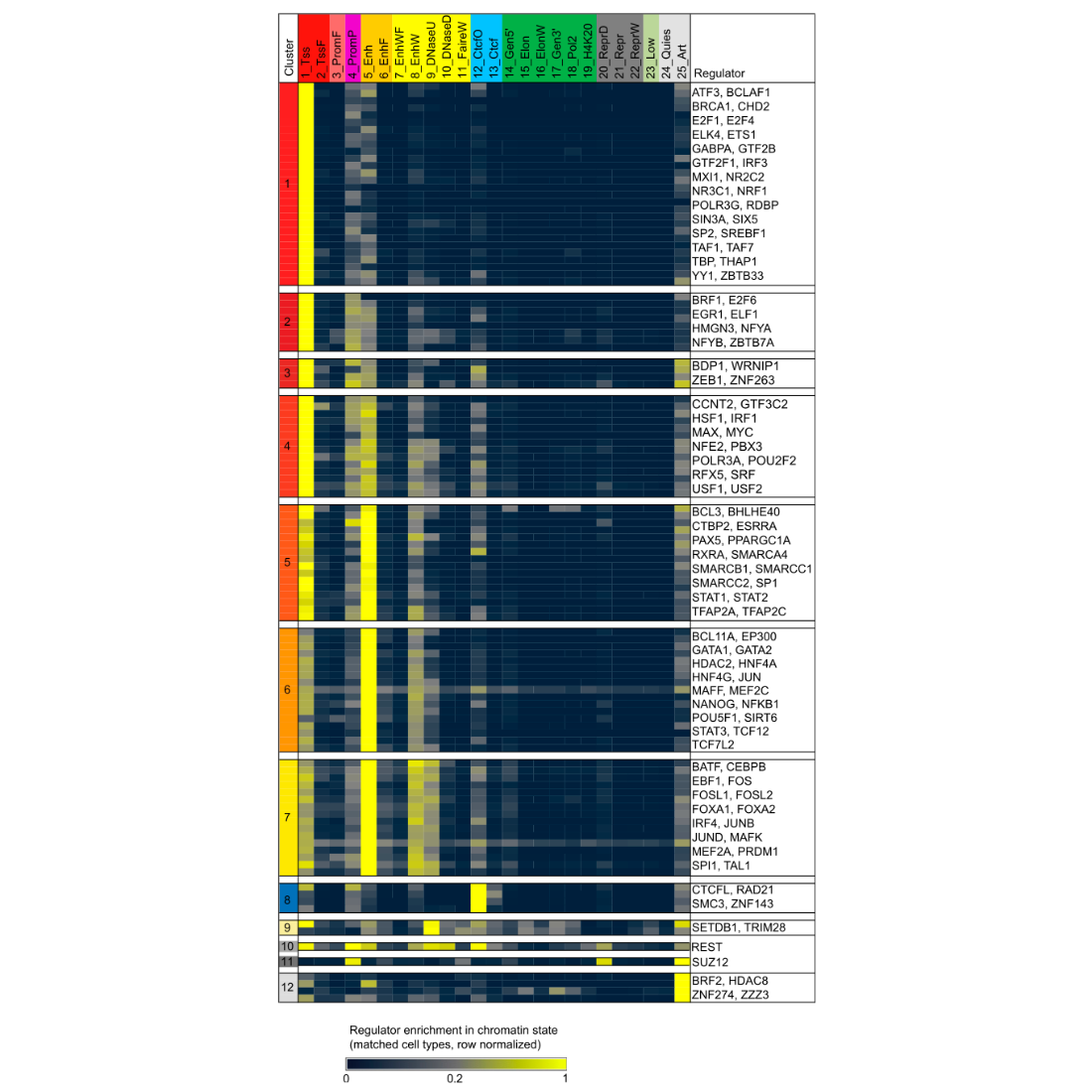 |
96.
Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. (pdf)
Journal: Genome Research, 2013 Jul doi: 10.1101/gr.144840.112 PMID:23595227 PMCID: PMC3698507 |
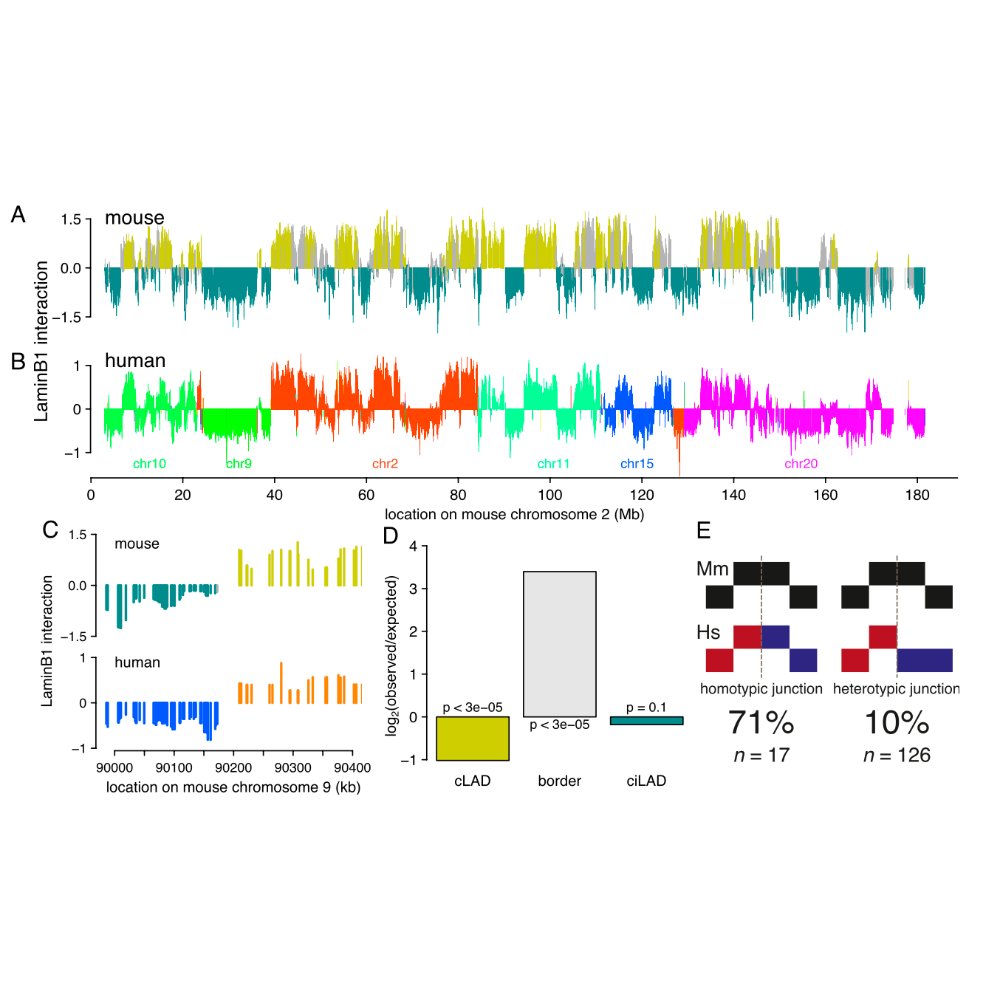 |
88.
Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. (pdf)
Journal: Genome Research, 2013 Feb doi: 10.1101/gr.141028.112 PMID:23124521 PMCID: PMC3561868 |
 |
85.
Integrative annotation of chromatin elements from ENCODE data. (pdf)
Journal: Nucleic Acids Research, 2013 Jan doi: 10.1093/nar/gks1284 PMID:23221638 PMCID: PMC3553955 |
 |
83.
RFECS: a random-forest based algorithm for enhancer identification from chromatin state. (pdf)
Journal: Plos Computational Biology, 2013 doi: 10.1371/journal.pcbi.1002968 PMID:23526891 PMCID: PMC3597546 |
 |
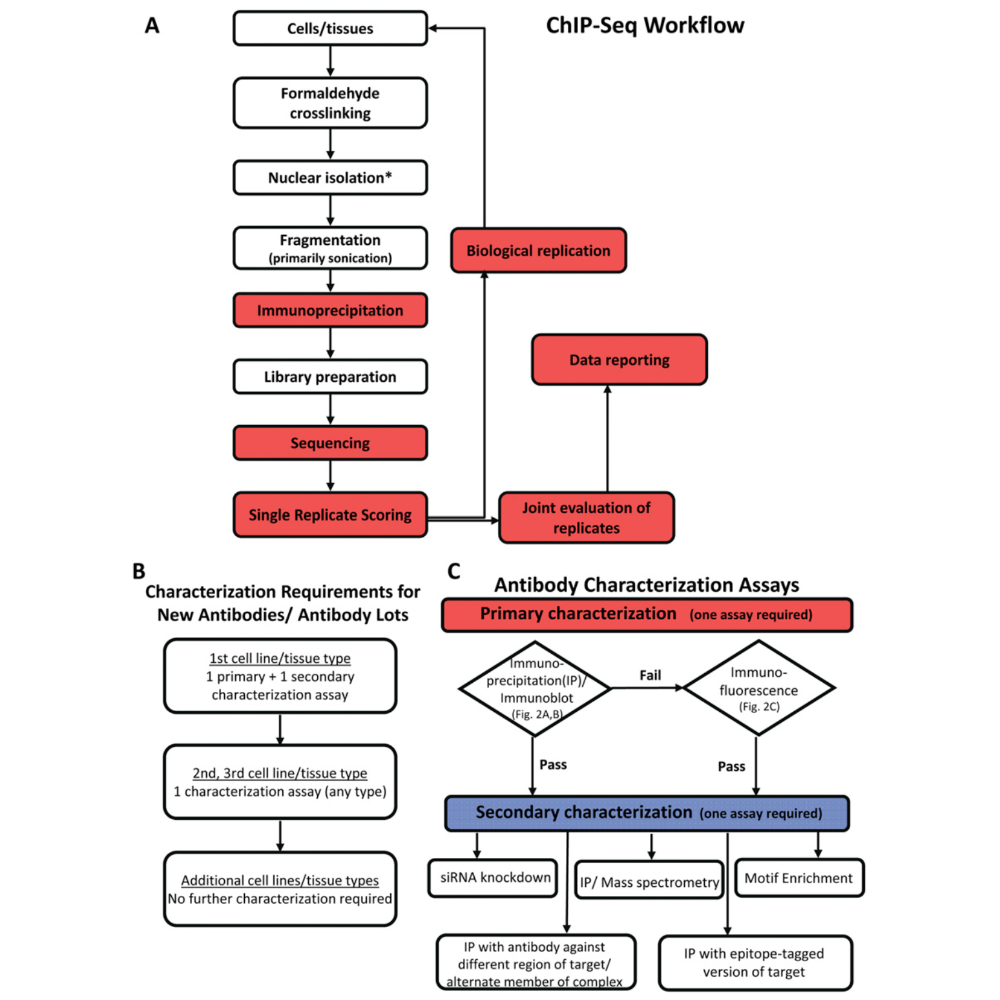
81.
Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. (pdf)
Journal: Cell Reports, 2012 Nov 29 doi: 10.1016/j.celrep.2012.10.017 PMID:23177624 PMCID: PMC3625945 |
 |
73.
ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. (pdf)
Journal: Genome Research, 2012 Sep doi: 10.1101/gr.136184.111 PMID:22955991 PMCID: PMC3431496 |
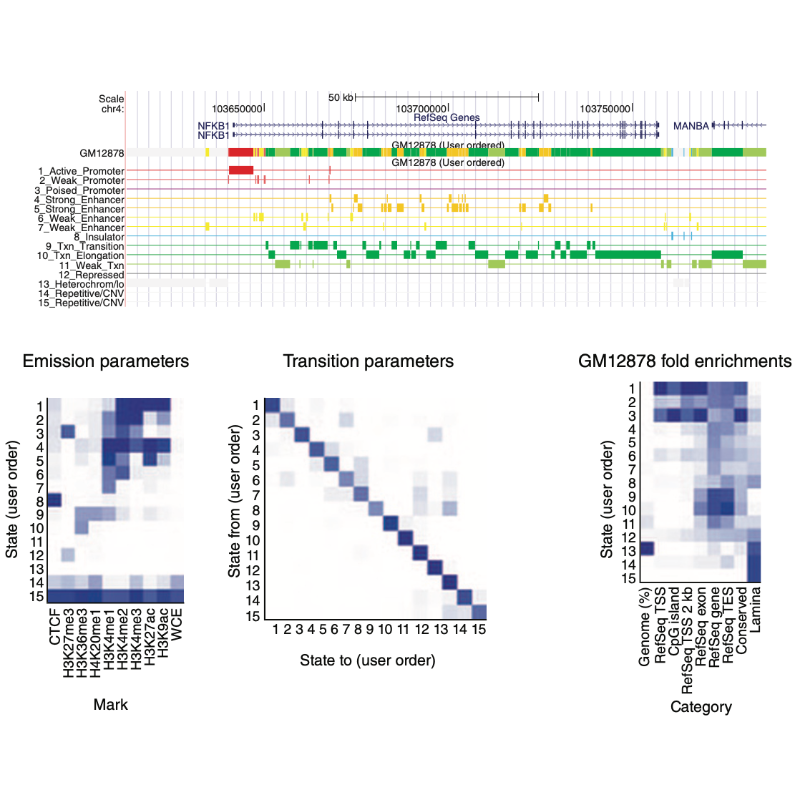 |
67.
ChromHMM: automating chromatin-state discovery and characterization. (pdf)
Journal: Nature Methods, 2012 Feb 28 doi: 10.1038/nmeth.1906 PMID:22373907 PMCID: PMC3577932 |
 |
61.
Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. (pdf)
Journal: Cell, 2011 Dec 23 doi: 10.1016/j.cell.2011.09.057 PMID:22196736 PMCID: PMC3312319 |
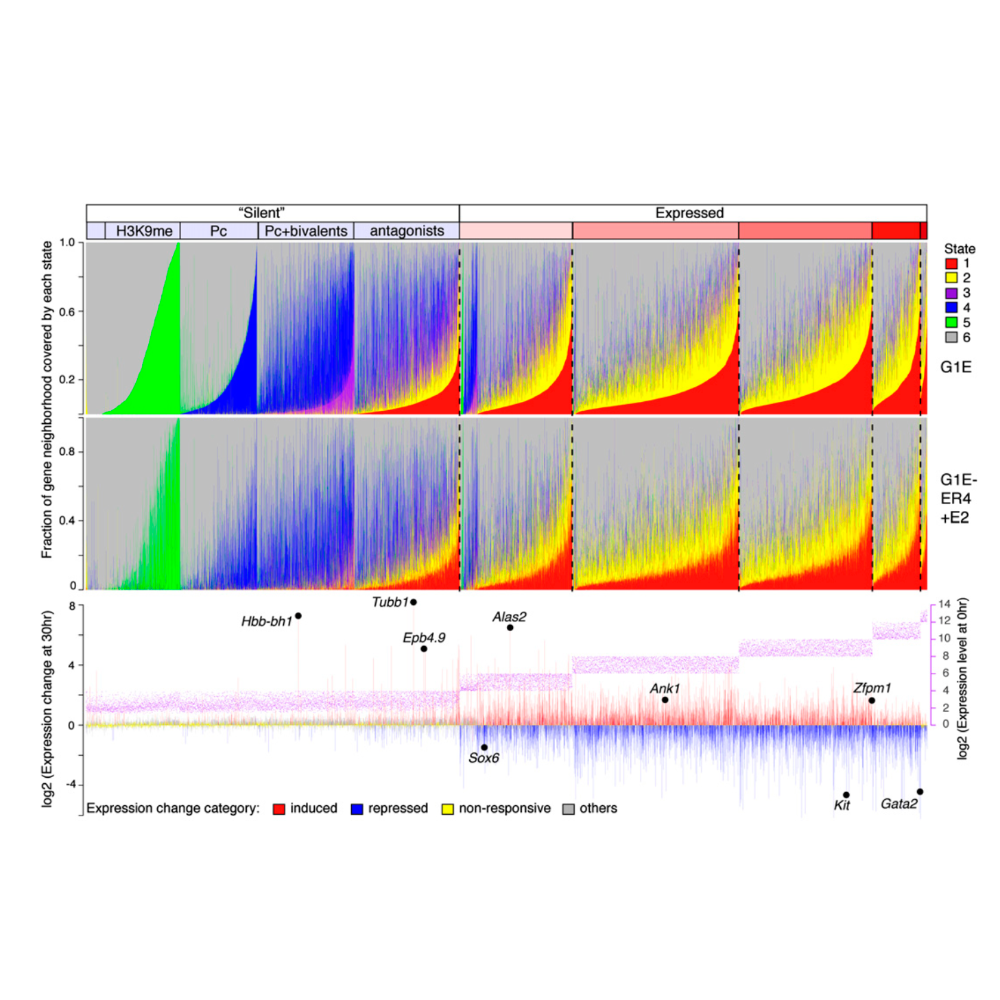 |
56.
Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. (pdf)
Journal: Genome Research, 2011 Oct doi: 10.1101/gr.125088.111 PMID:21795386 PMCID: PMC3202283 |
 |
51.
An epigenetic signature for monoallelic olfactory receptor expression. (pdf)
Journal: Cell, 2011 May 13 doi: 10.1016/j.cell.2011.03.040 PMID:21529909 PMCID: PMC3094500 |
 |
50.
Mapping and analysis of chromatin state dynamics in nine human cell types. (pdf)
Journal: Nature, 2011 May 5 doi: 10.1038/nature09906 PMID:21441907 PMCID: PMC3088773 |
 |
48.
Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. (pdf)
Journal: Nature, 2011 Mar 24 doi: 10.1038/nature09725 PMID:21179089 PMCID: PMC3109908 |
 |
40.
The NIH Roadmap Epigenomics Mapping Consortium. (pdf)
Journal: Nature Biotechnology, 2010 Oct doi: 10.1038/nbt1010-1045 PMID:20944595 PMCID: PMC3607281 |
 |
39.
Discovery and characterization of chromatin states for systematic annotation of the human genome. (pdf)
Journal: Nature Biotechnology, 2010 Aug doi: 10.1038/nbt.1662 PMID:20657582 PMCID: PMC2919626 |
 |
38.
A comprehensive map of insulator elements for the Drosophila genome. (pdf)
Journal: Plos Genetics, 2010 Jan 15 doi: 10.1371/journal.pgen.1000814 PMID:20084099 PMCID: PMC2797089 |
 |
31.
Histone modifications at human enhancers reflect global cell-type-specific gene expression. (pdf)
Journal: Nature, 2009 May 7 doi: 10.1038/nature07829 PMID:19295514 PMCID: PMC2910248 |
 |
21.
RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. (pdf)
Journal: Nature Genetics, 2007 Dec doi: 10.1038/ng.2007.26 PMID:17994019 PMCID: PMC2824921 |
 |
12.
Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. (pdf)
Journal: Genes & Development, 2007 Feb 15 doi: 10.1101/gad.1509607 PMID:17322397 PMCID: PMC1804326 |
 |
7.
Transcriptional regulatory code of a eukaryotic genome. (pdf)
Journal: Nature, 2004 Sep 2 doi: 10.1038/nature02800 PMID:15343339 PMCID: PMC3006441 |
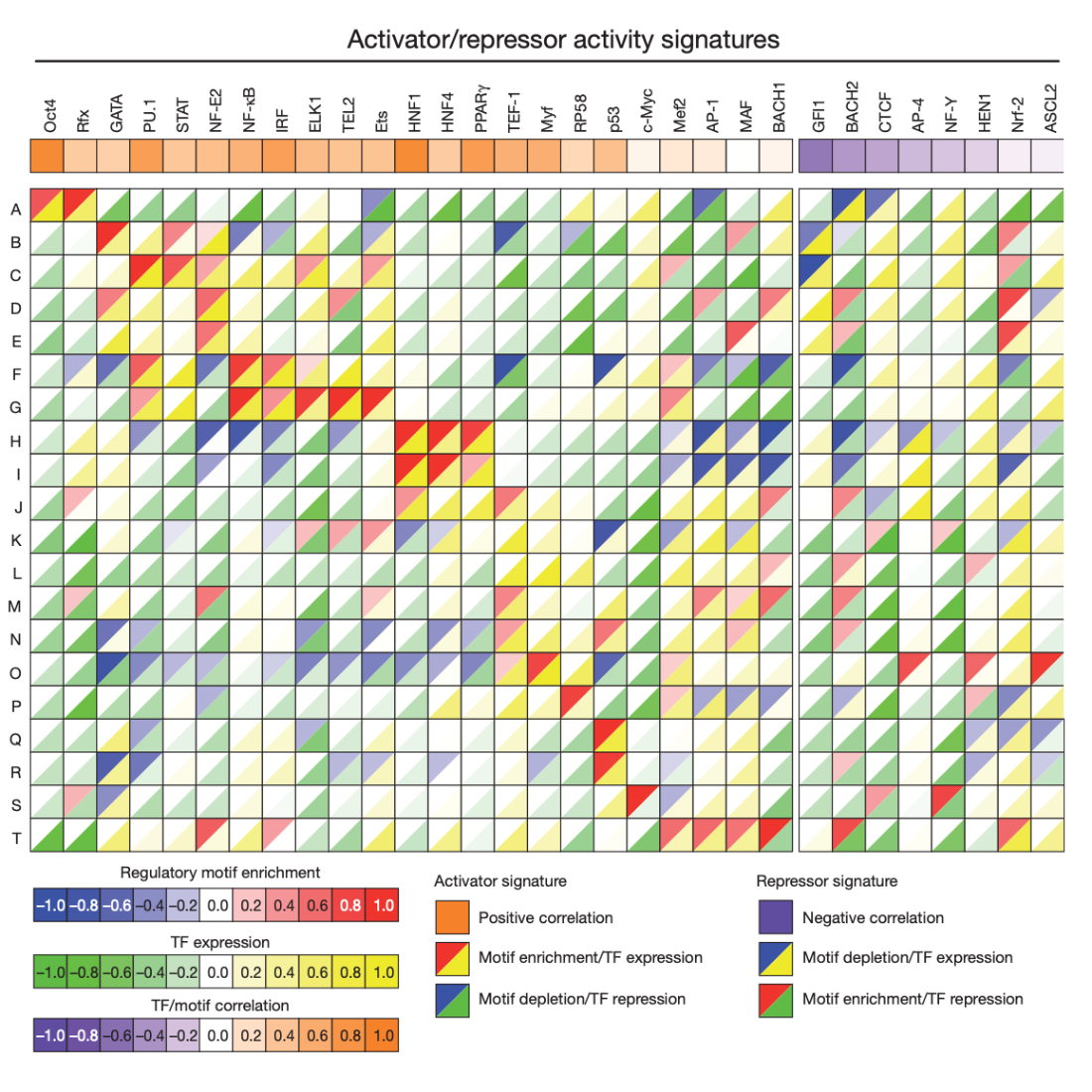
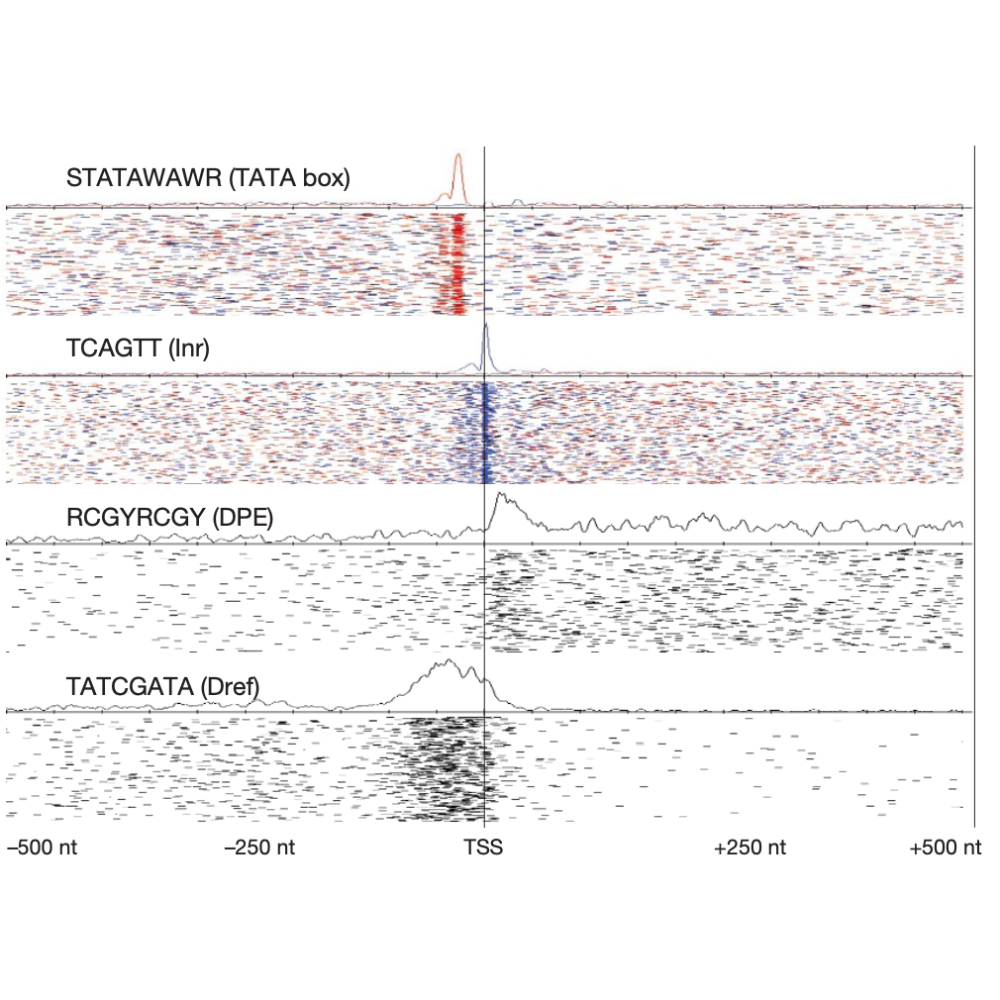
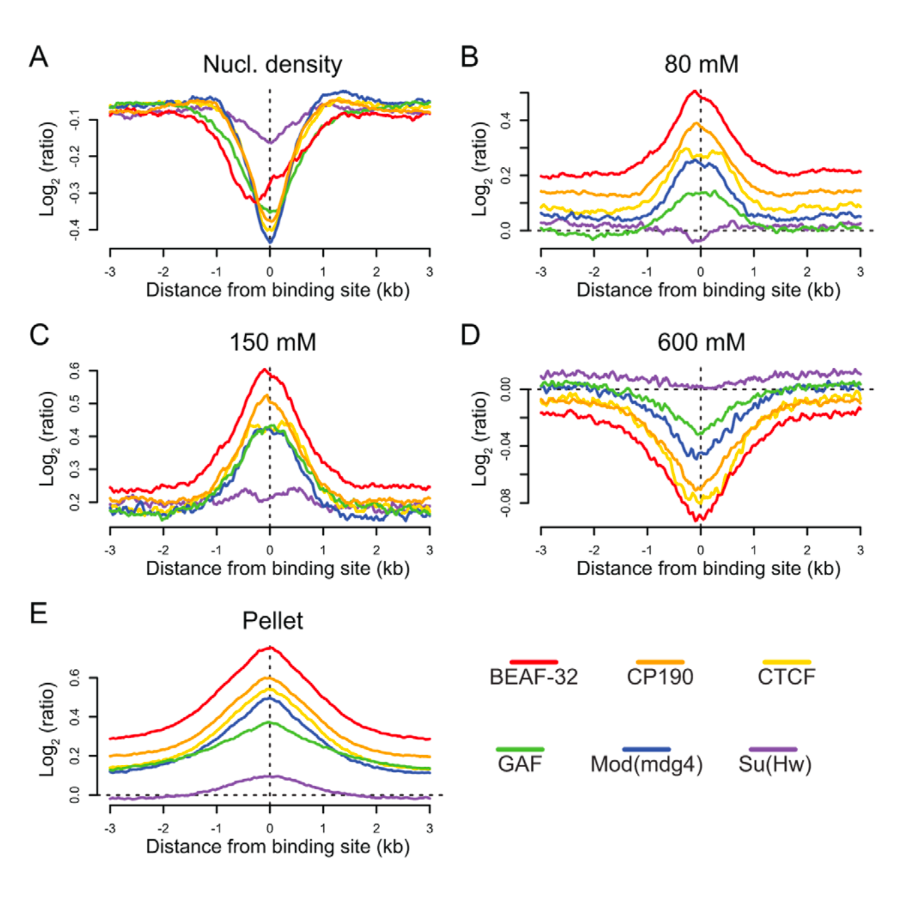
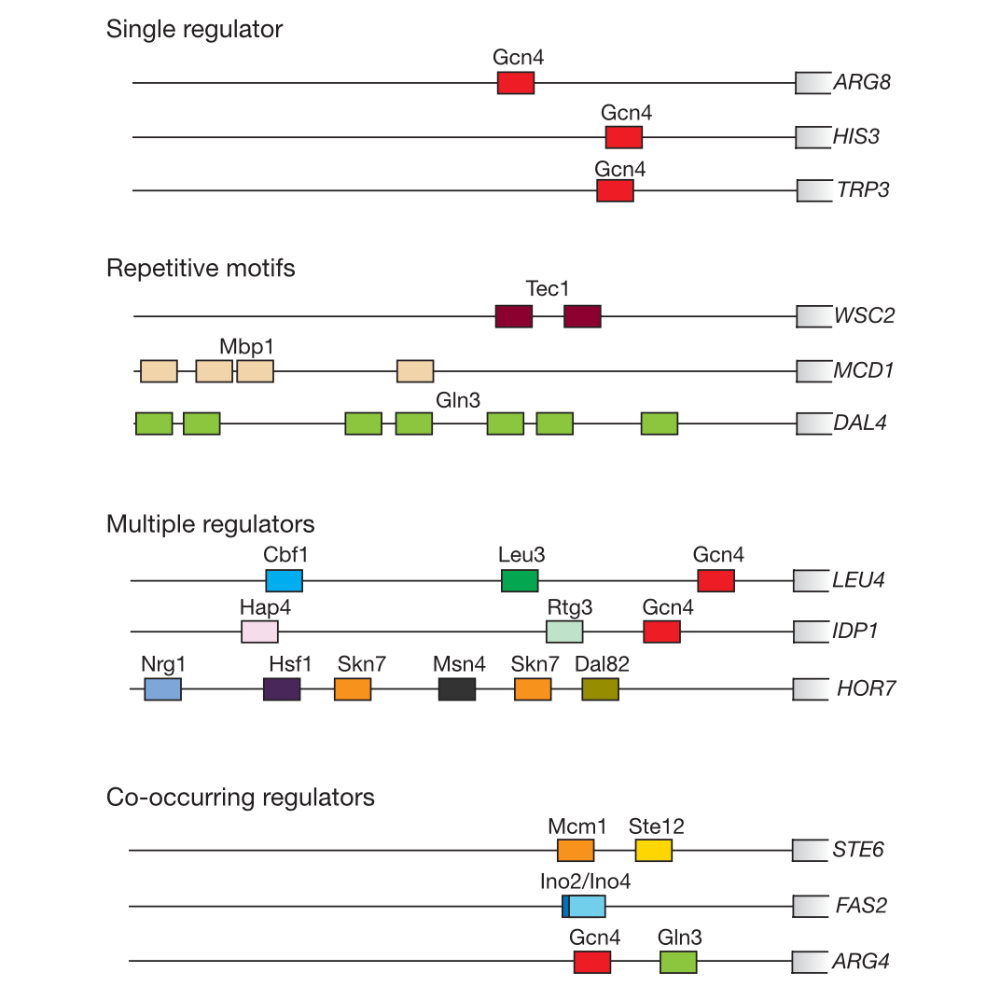
Regulatory Motifs
Beyond a comprehensive encyclopedia of genomic elements we need to understand how these elements are put together in cellular circuits and to infer predictive models of cellular response, which can serve as foundations for modern medicine, rational drug design, and personalized treatments. Cellular regulatory networks that control gene expression are dictated by the binding of DNA and RNA regulators to sequence motifs typically only 6-8 nucleotides long. These 'regulatory motifs' have previously remained elusive to systematic discovery due to their short lengths and varying distances at which they can act. However, by evaluating their genome-wide conservation patterns, their positional and strand biases, and their association with functional genomic datasets, our work has pioneered new methods for their systematic discovery, the characterization of their functional roles, and the inference of the regulatory networks they define. We also mined time-course and multi-cell type datasets to reveal the stage- and tissue-specific action of the regulators that recognize these motifs, and to build computational models that predict gene expression from the combined action of multiple regulators for each gene.
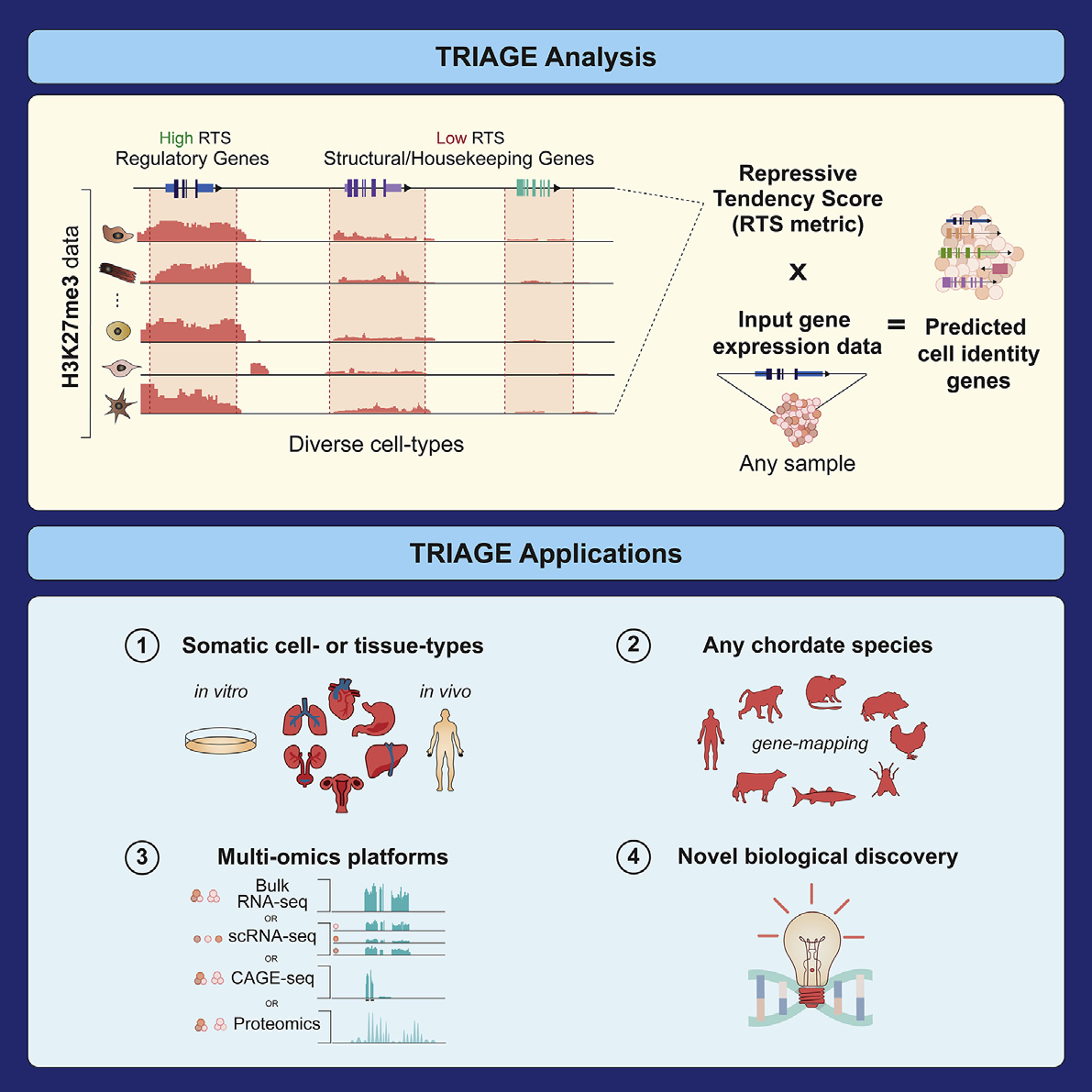 |
264.
Conserved Epigenetic Regulatory Logic Infers Genes Governing Cell Identity. (pdf)
Journal: Cell Systems, 2020 Dec 16 doi: 10.1016/j.cels.2020.11.001 PMID:33278344 PMCID: PMC7781436 |
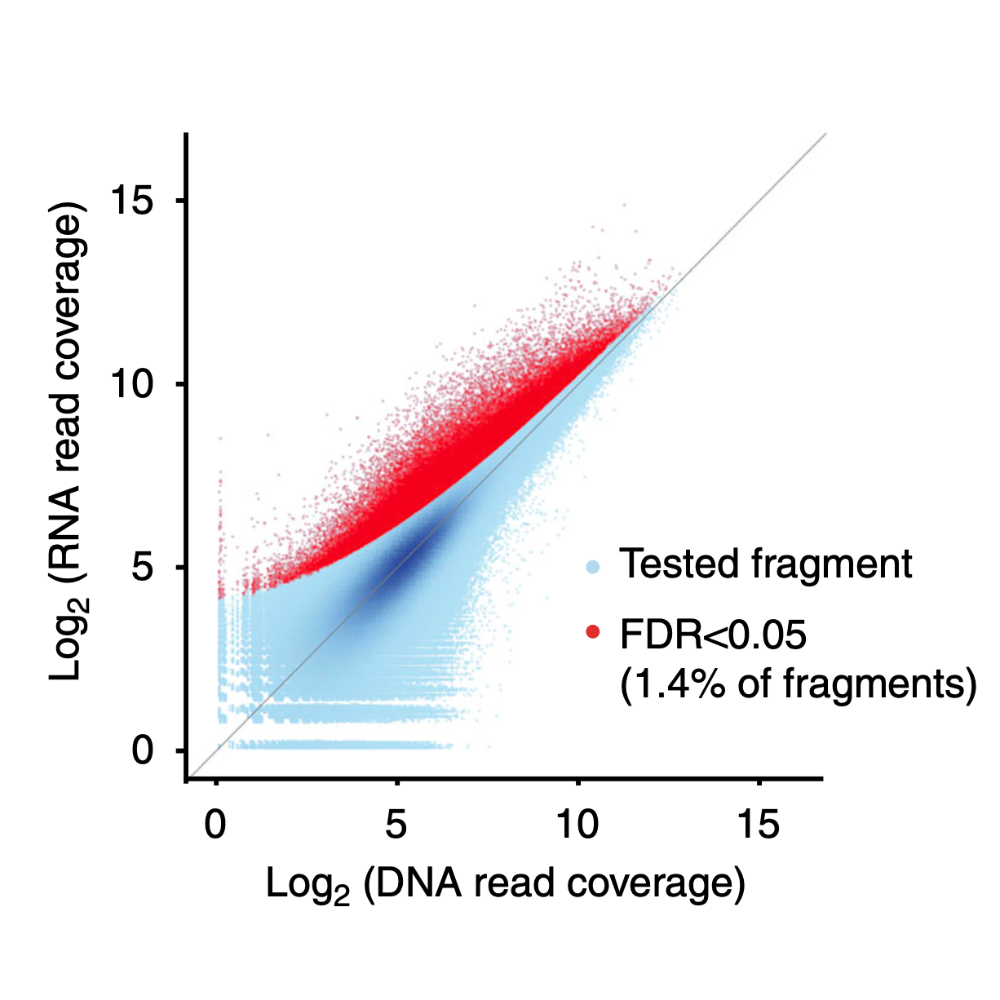 |
195.
High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human. (pdf)
Journal: Nature Communications, 2018 Dec 19 doi: 10.1038/s41467-018-07746-1 PMID:30568279 PMCID: PMC6300699 |
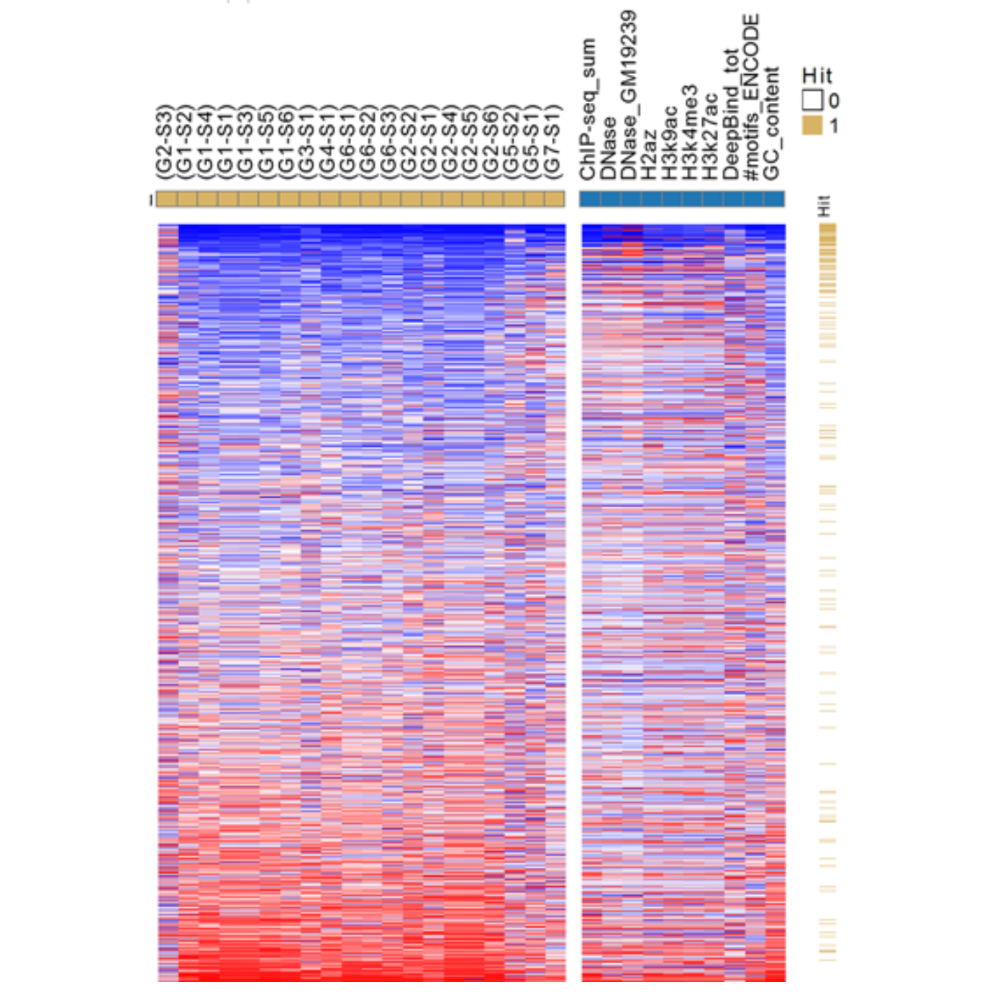 |
169.
Predicting gene expression in massively parallel reporter assays: A comparative study. (pdf)
Journal: Human Mutation, 2017 Sep doi: 10.1002/humu.23197 PMID:28220625 PMCID: PMC5560998 |
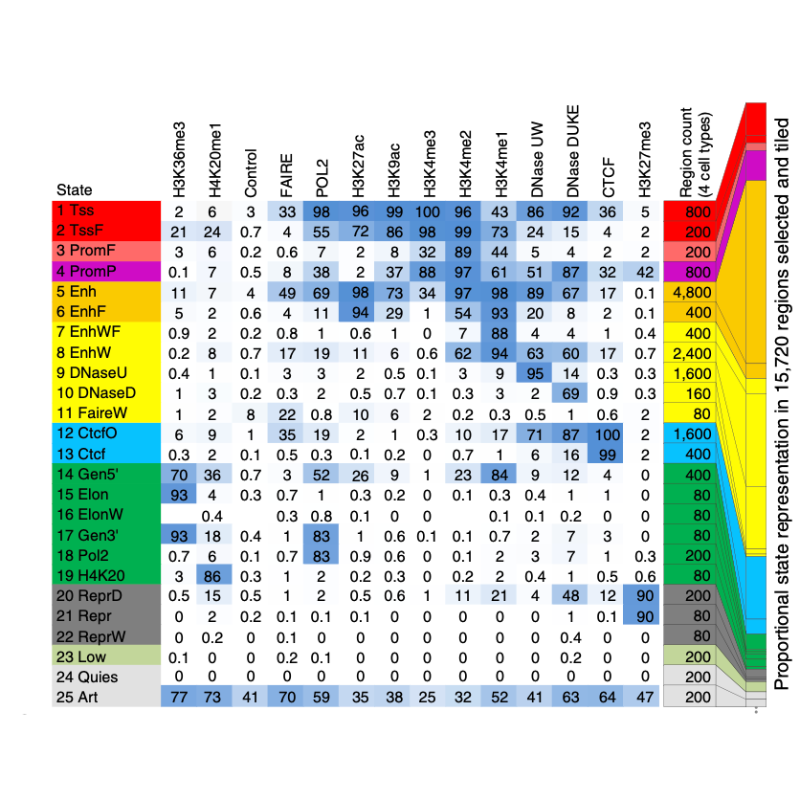 |
162.
Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. (pdf)
Journal: Nature Biotechnology, 2016 Nov doi: 10.1038/nbt.3678 PMID:27701403 PMCID: PMC5125825 |
 |
155.
Survey of variation in human transcription factors reveals prevalent DNA binding changes. (pdf)
Journal: Science, 2016 Mar 25 doi: 10.1126/science.aad2257 PMID:27013732 PMCID: PMC4825693 |
 |
146.
Deep learning for regulatory genomics. (pdf)
Journal: Nature Biotechnology, 2015 Aug doi: 10.1038/nbt.3313 PMID:26252139 |
 |
144.
Context influences on TALE-DNA binding revealed by quantitative profiling. (pdf)
Journal: Nature Communications, 2015 Jun 11 doi: 10.1038/ncomms8440 PMID:26067805 PMCID: PMC4467457 |
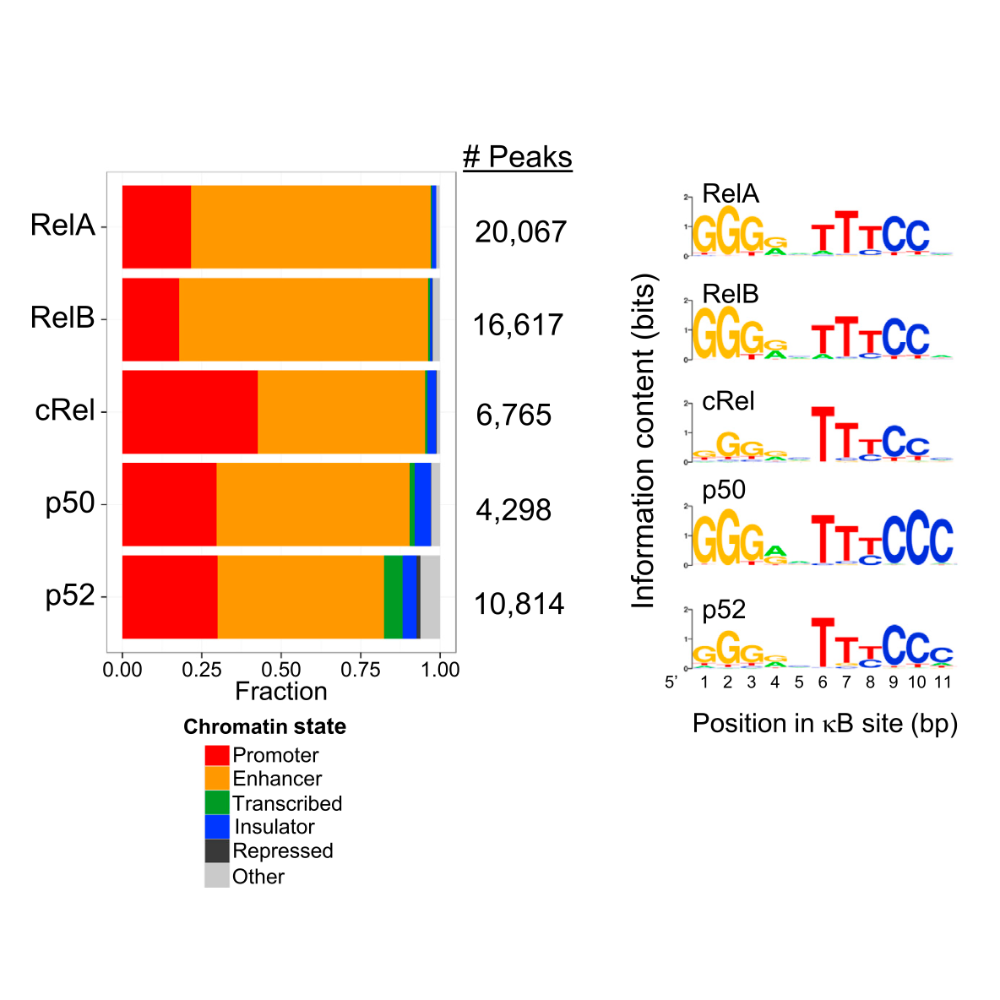 |
124.
The NF-kappaB genomic landscape in lymphoblastoid B cells. (pdf)
Journal: Cell Reports, 2014 Sep 11 doi: 10.1016/j.celrep.2014.07.037 PMID:25159142 PMCID: PMC4163118 |
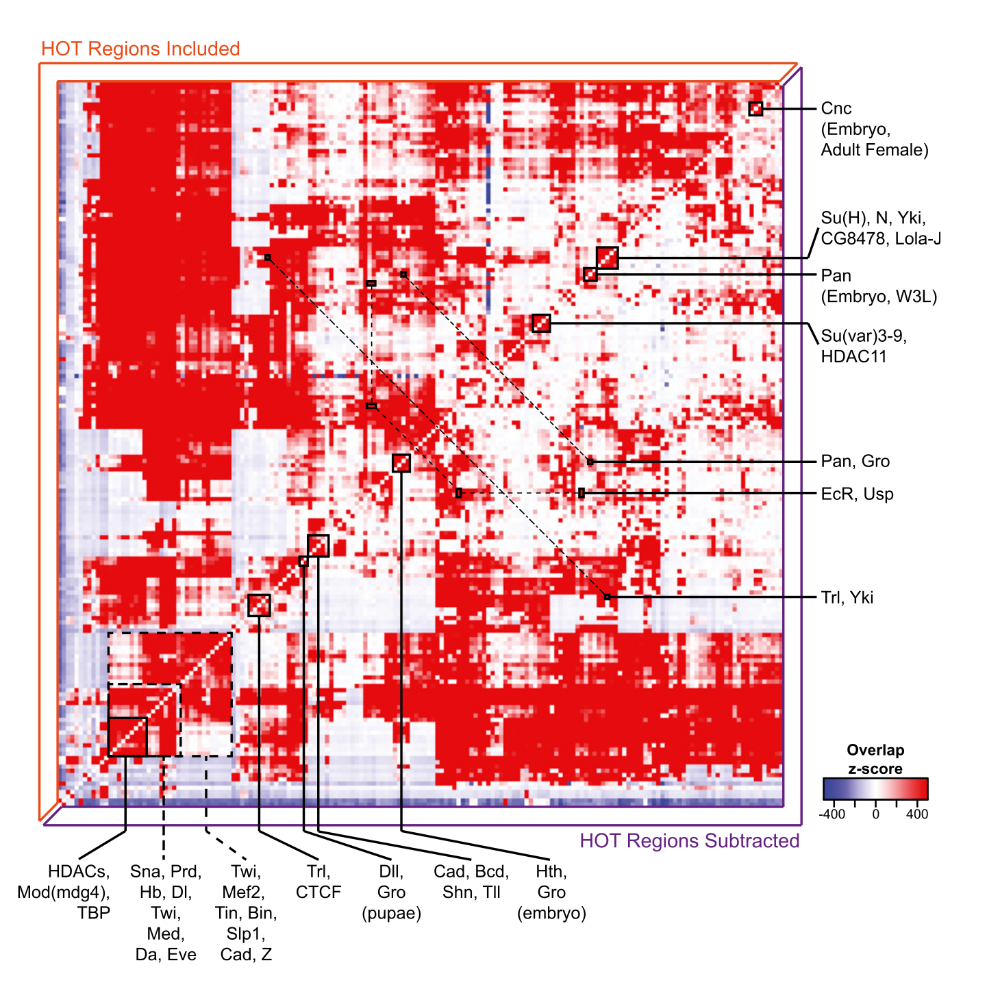 |
116.
Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster. (pdf)
Journal: Genome Research, 2014 Jul doi: 10.1101/gr.168807.113 PMID:24985916 PMCID: PMC4079976 |
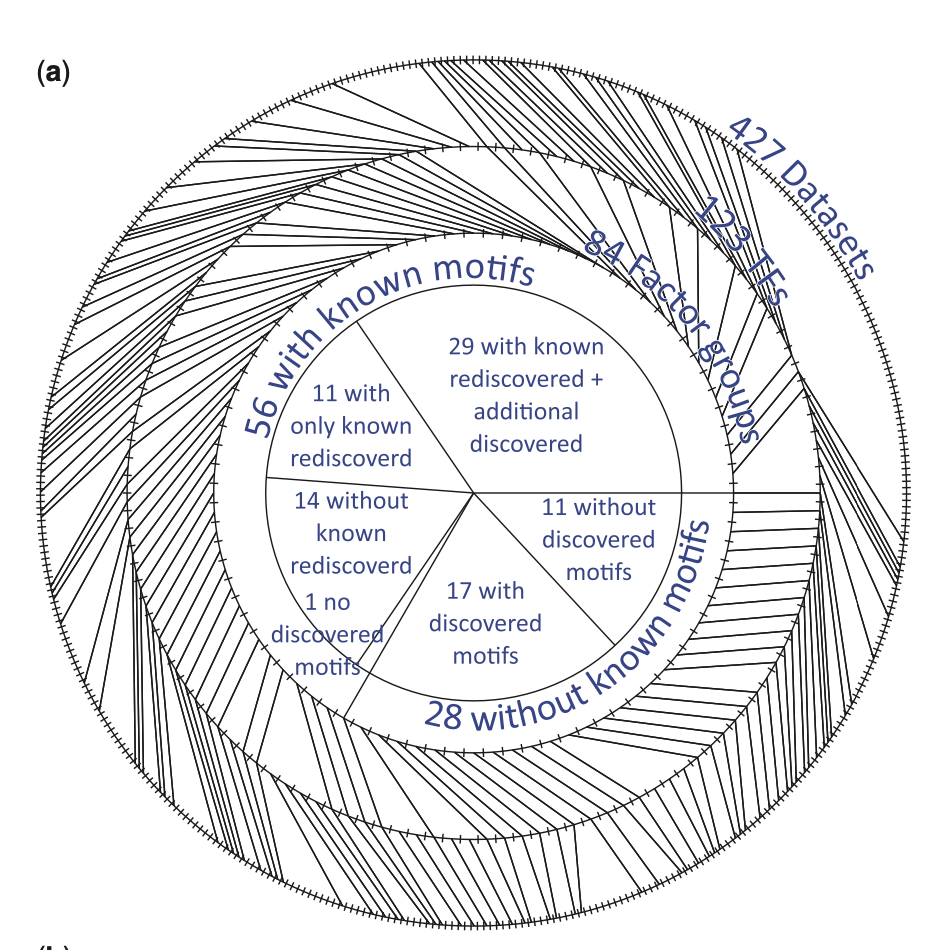 |
108.
Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. (pdf)
Journal: Nucleic Acids Research, 2014 Mar doi: 10.1093/nar/gkt1249 PMID:24335146 PMCID: PMC3950668 |
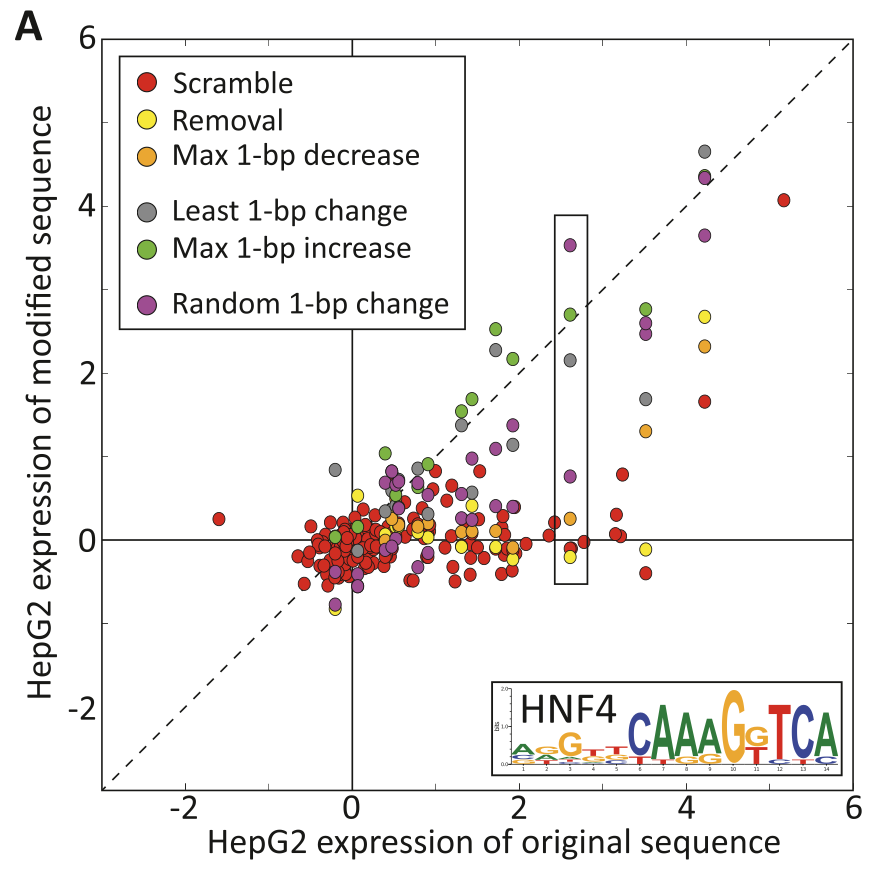 |
91.
Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. (pdf)
Journal: Genome Research, 2013 May doi: 10.1101/gr.144899.112 PMID:23512712 PMCID: PMC3638136 |
 |
78.
Analysis of variation at transcription factor binding sites in Drosophila and humans. (pdf)
Journal: Genome Biology, 2012 Sep 28 doi: 10.1186/gb-2012-13-9-r49 PMID:22950968 PMCID: PMC3491393 |
 |
66.
Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. (pdf)
Journal: Nature Biotechnology, 2012 Feb 26 doi: 10.1038/nbt.2137 PMID:22371084 PMCID: PMC3297981 |
 |
47.
A cis-regulatory map of the Drosophila genome. (pdf)
Journal: Nature, 2011 Mar 24 doi: 10.1038/nature09990 PMID:21430782 PMCID: PMC3179250 |
 |
36.
Motif Discovery in Physiological Datasets: A Methodology for Inferring Predictive Elements. (pdf)
Journal: Acm Transactions On Knowledge Discovery From Data, 2010 Jan doi: 10.1145/1644873.1644875 PMID:20730037 PMCID: PMC2923403 |
 |
10.
Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. (pdf)
Journal: Nature, 2005 Mar 17 doi: 10.1038/nature03441 PMID:15735639 PMCID: PMC2923337 |
 |
3.
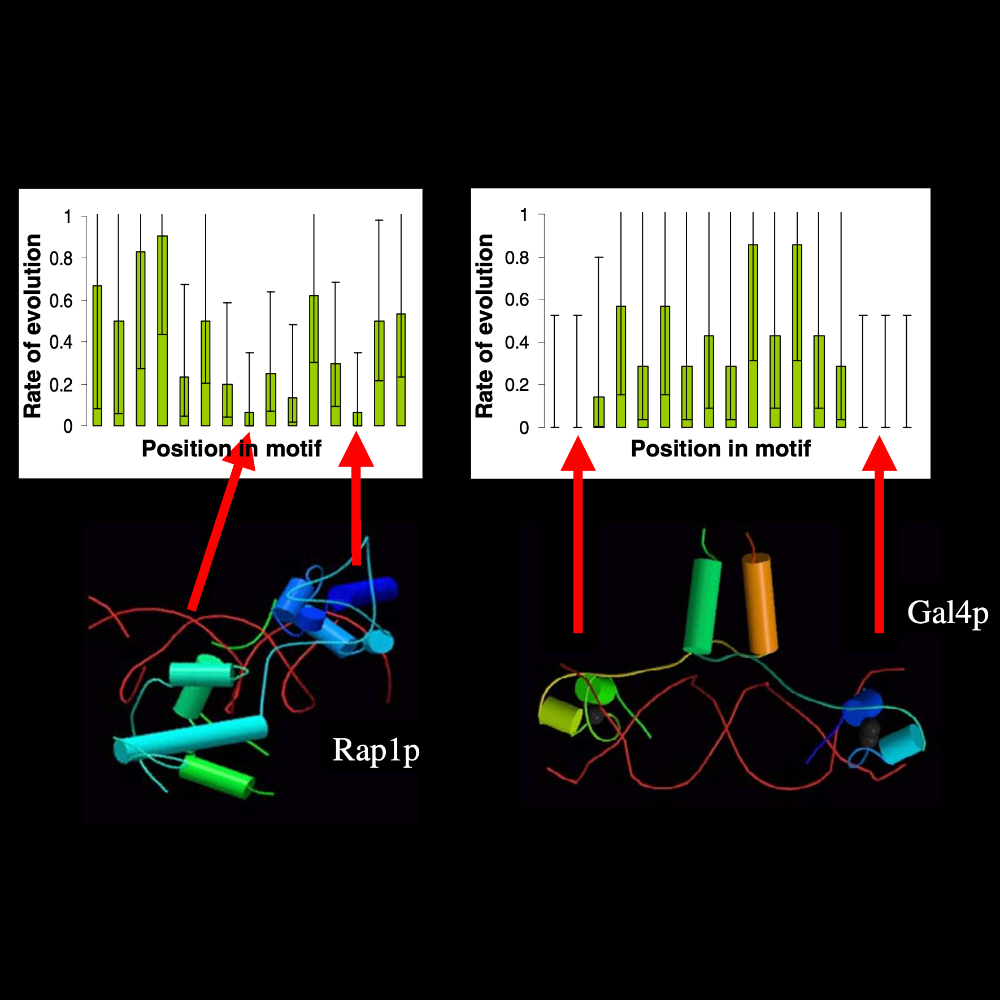
Position specific variation in the rate of evolution in transcription factor binding sites. (pdf)
Journal: Bmc Evolutionary Biology, 2003 Aug 28 doi: 10.1186/1471-2148-3-19 PMID:12946282 PMCID: PMC212491 |
 |
2.
Sequencing and comparison of yeast species to identify genes and regulatory elements. (pdf)
Journal: Nature, 2003 May 15 doi: 10.1038/nature01644 PMID:12748633 |
 |
1.
Phylogenetically and spatially conserved word pairs associated with gene-expression changes in yeasts. (pdf)
Journal: Genome Biology, 2003 doi: 10.1186/gb-2003-4-7-r43 PMID:12844359 PMCID: PMC193630 |
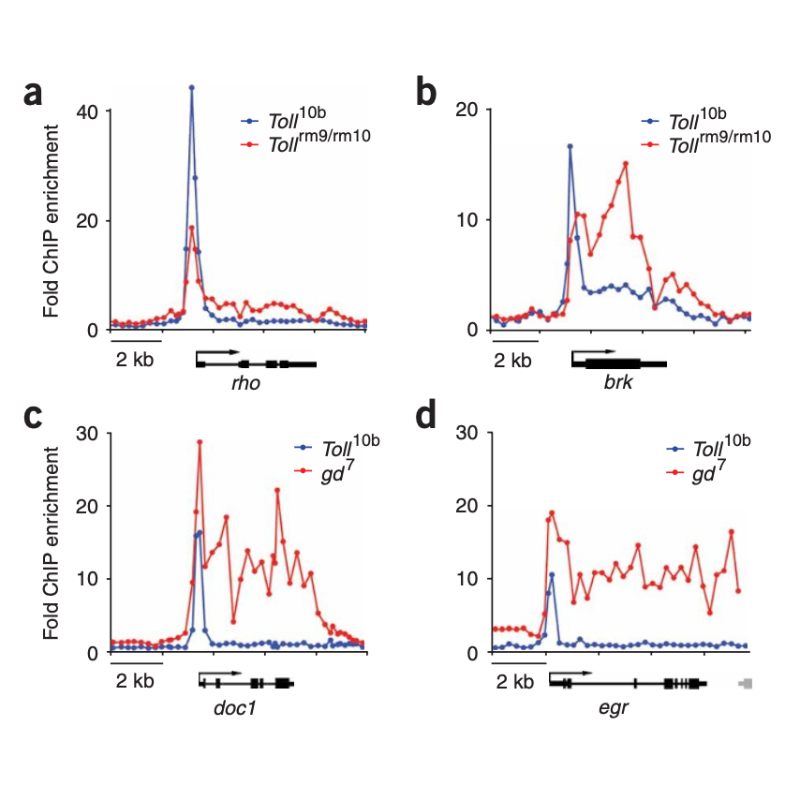
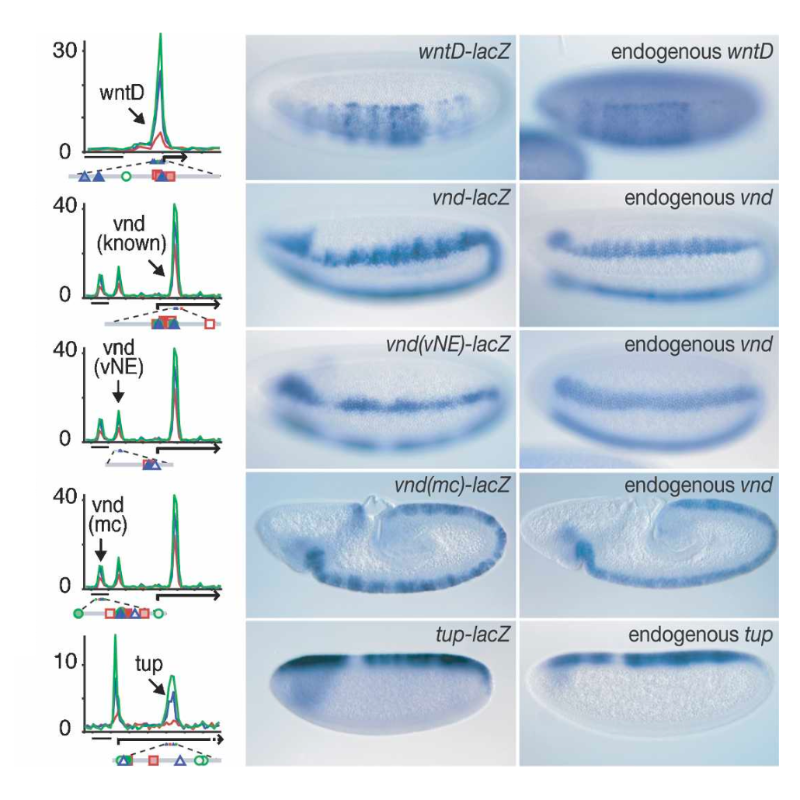
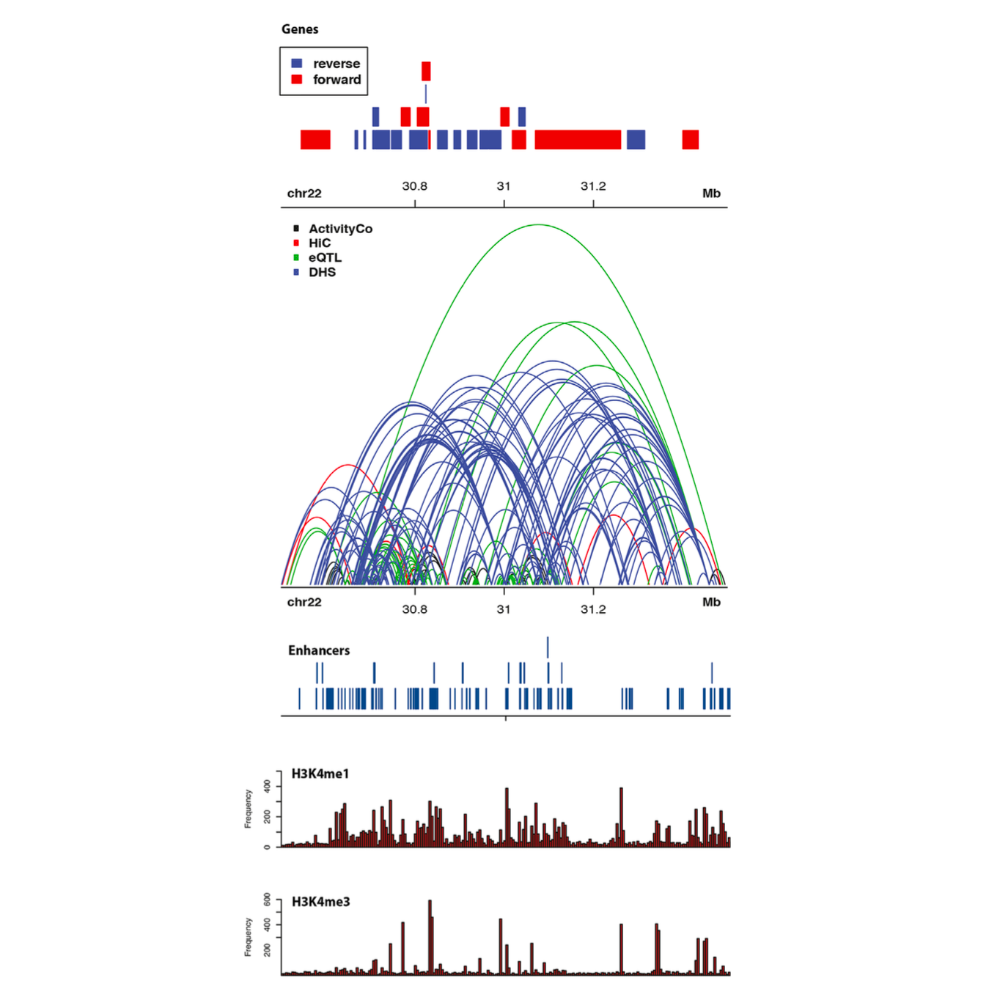
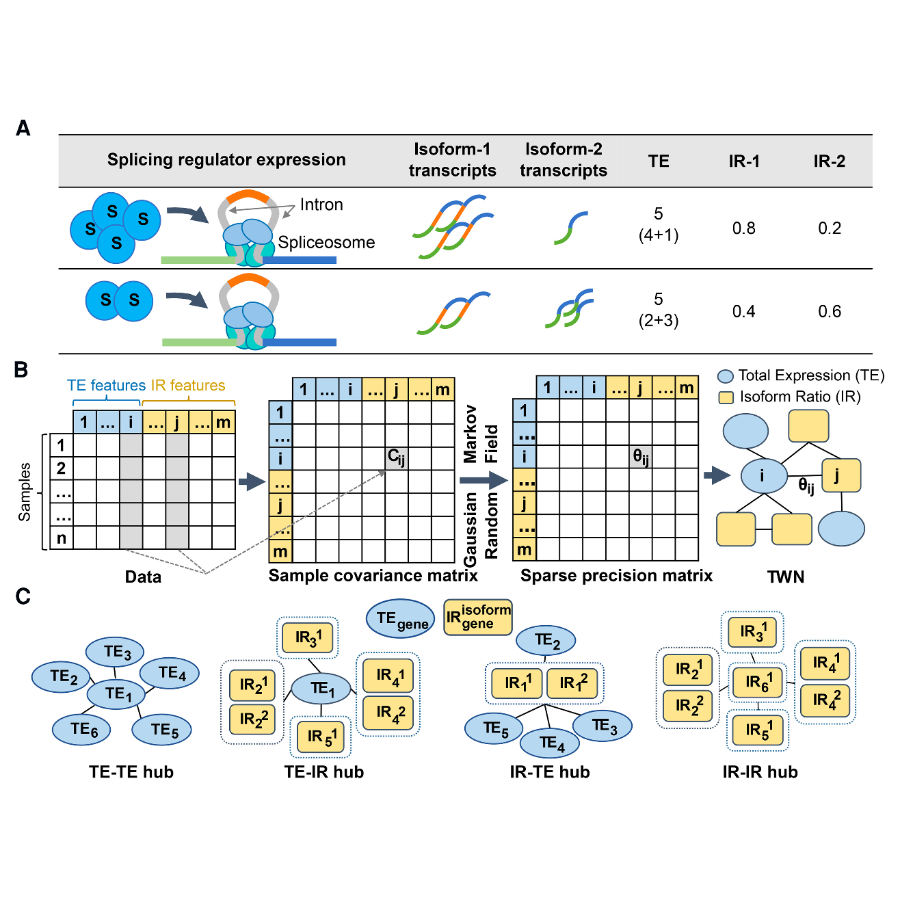
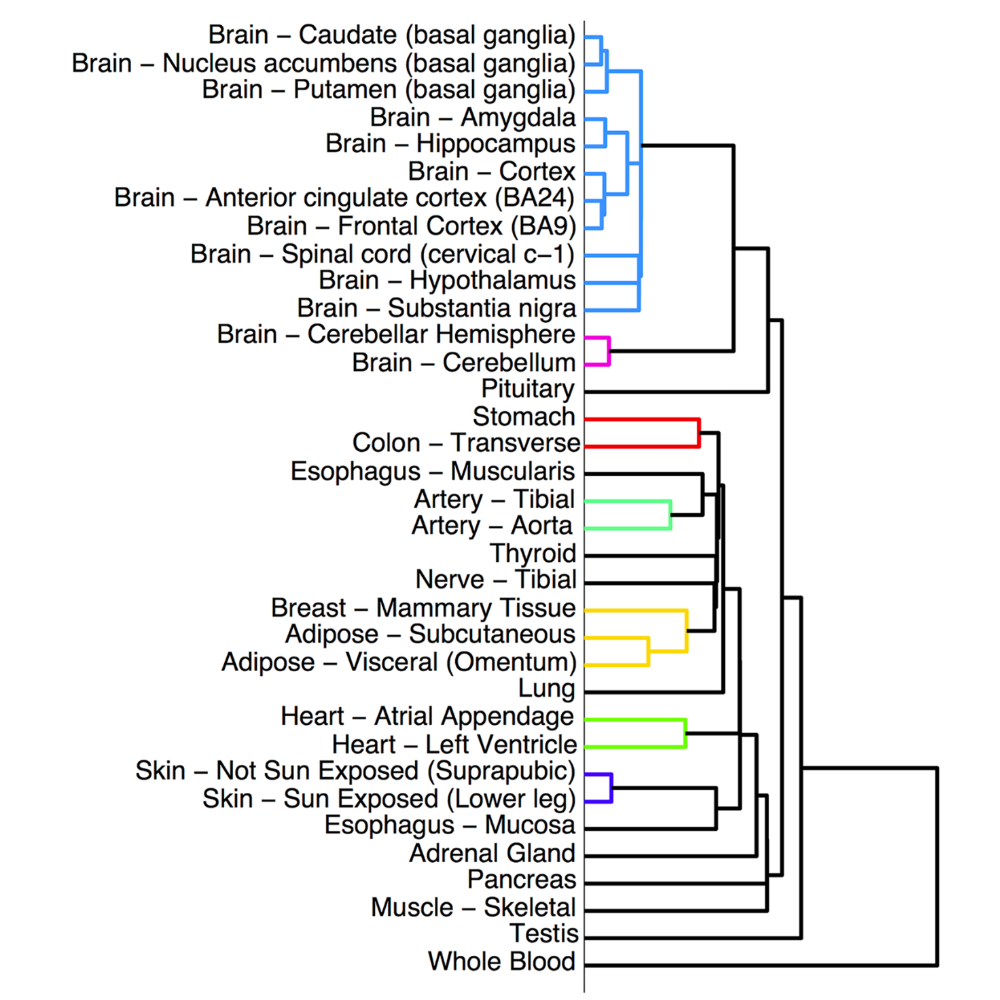
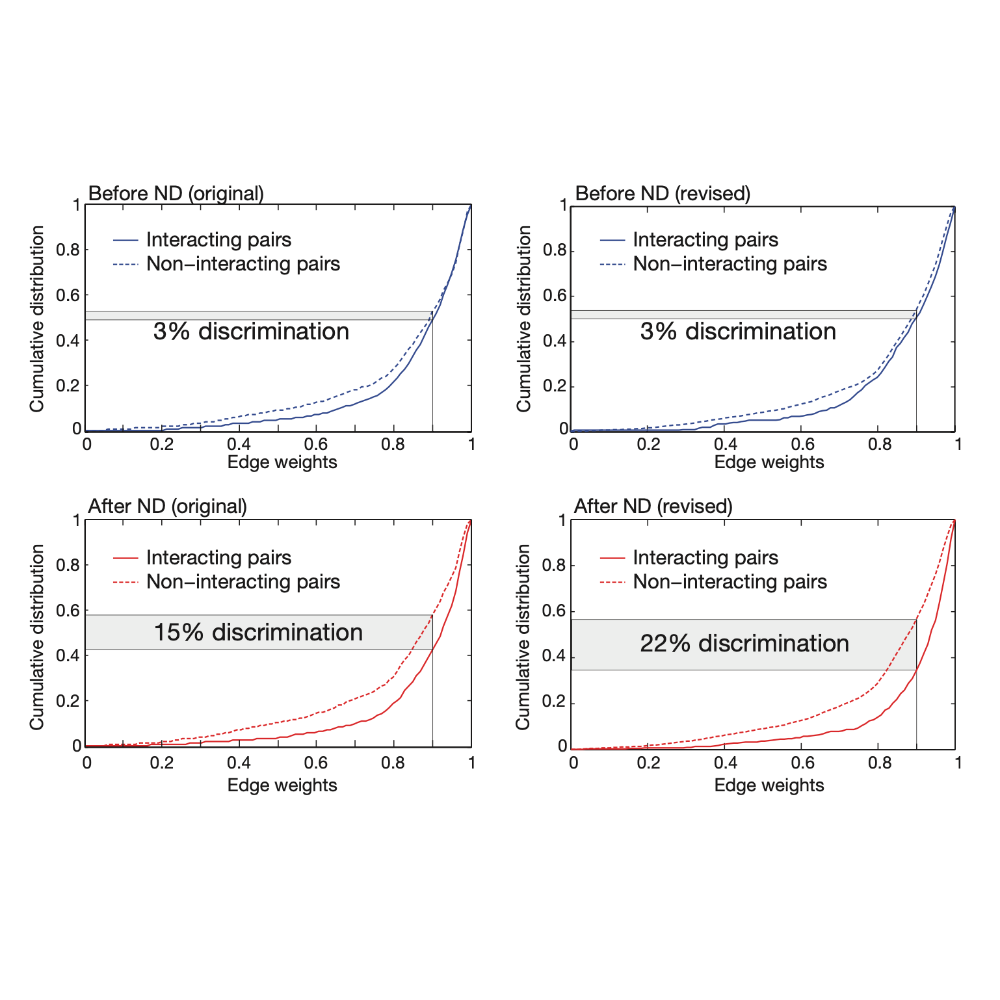
Regulatory Networks
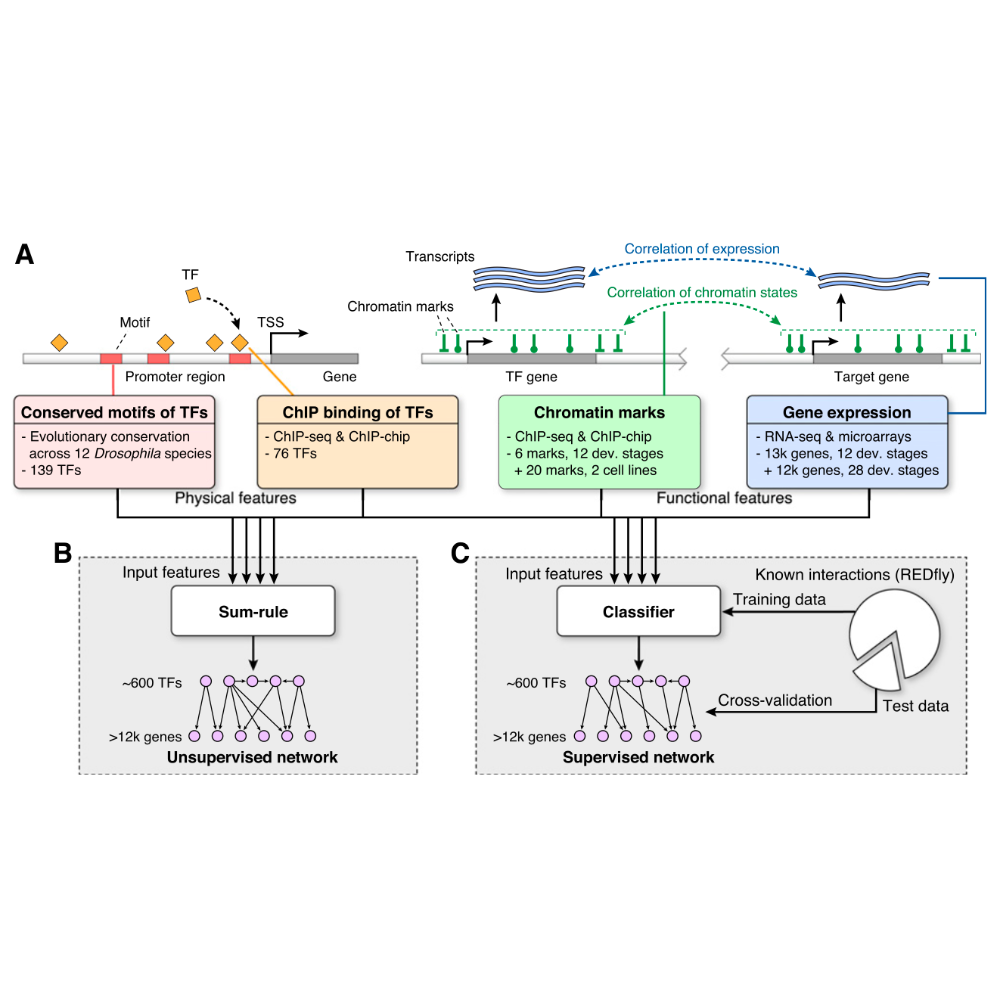
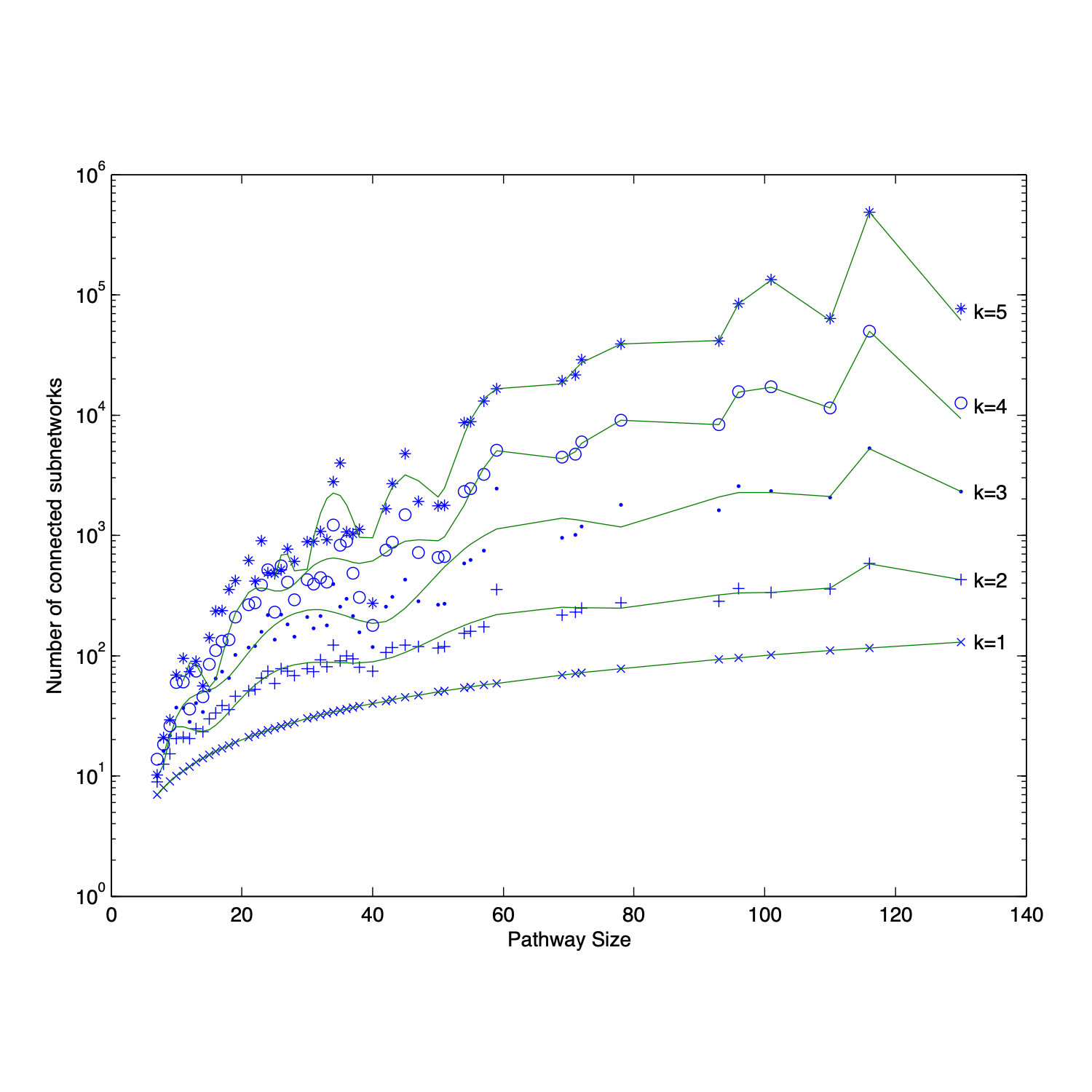
While the genome is static and its code one-dimentional, the binding of regulators to target genes, which can themselves be regulators of other targets, defines complex circuits of information flow, or 'regulatory networks'. However, defining these networks can be a major challenge given the complex and sometimes subtle relationships between regulators and their targets. To address these challenges, we developed methods for de novo discovery of regulator targets, using a phylogenetic framework for motif conservation, and rigorous statistics of motif confidence. We showed that computationally-derived regulator targets are of comparable quality and abundance as ChIP-derived binding sites (showing stronger overlap with target genes than ChIP, even outside ChIP-bound regions), and that miRNA targets are of even higher confidence. We presented the first regulatory network for a complete animal genome and studied its structural and functional properties. Combining evolutionarily conserved motif instances with chromatin binding and expression analysis, we have defined functional regulatory networks for yeast under multiple stress conditions, and fly across developmental stages. We have also developed graph algorithms for discovering recurrent patterns of network connectivity known as 'network motifs' in both directed and undirected graphs, revealing combinatorial connectivity patterns in the transcriptional machinery.
 |
206.
Integrative construction of regulatory region networks in 127 human reference epigenomes by matrix factorization. (pdf)
Journal: Nucleic Acids Research, 2019 Aug 22 doi: 10.1093/nar/gkz538 PMID:31265076 PMCID: PMC6698807 |
 |
176.
Co-expression networks reveal the tissue-specific regulation of transcription and splicing. (pdf)
Journal: Genome Research, 2017 Nov doi: 10.1101/gr.216721.116 PMID:29021288 PMCID: PMC5668942 |
 |
156.
Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. (pdf)
Journal: Nature Methods, 2016 Apr doi: 10.1038/nmeth.3799 PMID:26950747 PMCID: PMC4967716 |
 |
155.
Survey of variation in human transcription factors reveals prevalent DNA binding changes. (pdf)
Journal: Science, 2016 Mar 25 doi: 10.1126/science.aad2257 PMID:27013732 PMCID: PMC4825693 |
 |
140.
Sharing and Specificity of Co-expression Networks across 35 Human Tissues. (pdf)
Journal: Plos Computational Biology, 2015 May doi: 10.1371/journal.pcbi.1004220 PMID:25970446 PMCID: PMC4430528 |
 |
137.
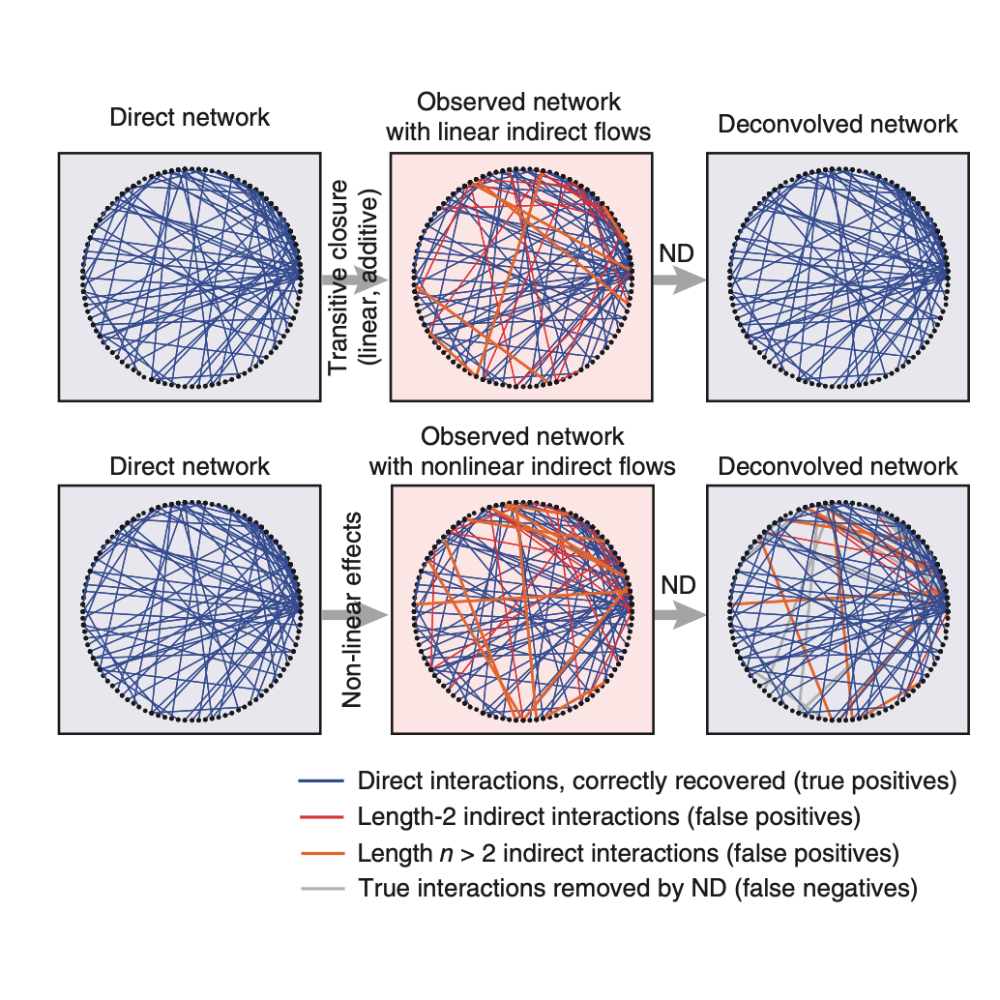
Corrigendum: Network deconvolution as a general method to distinguish direct dependencies in networks. (pdf)
Journal: Nature Biotechnology, 2015 Apr doi: 10.1038/nbt0415-424 PMID:25850062 |
 |
121.
Comparative analysis of regulatory information and circuits across distant species. (pdf)
Journal: Nature, 2014 Aug 28 doi: 10.1038/nature13668 PMID:25164757 PMCID: PMC4336544 |
 |
98.
Network deconvolution as a general method to distinguish direct dependencies in networks. (pdf)
Journal: Nature Biotechnology, 2013 Aug doi: 10.1038/nbt.2635 PMID:23851448 PMCID: PMC3773370 |
 |
72.
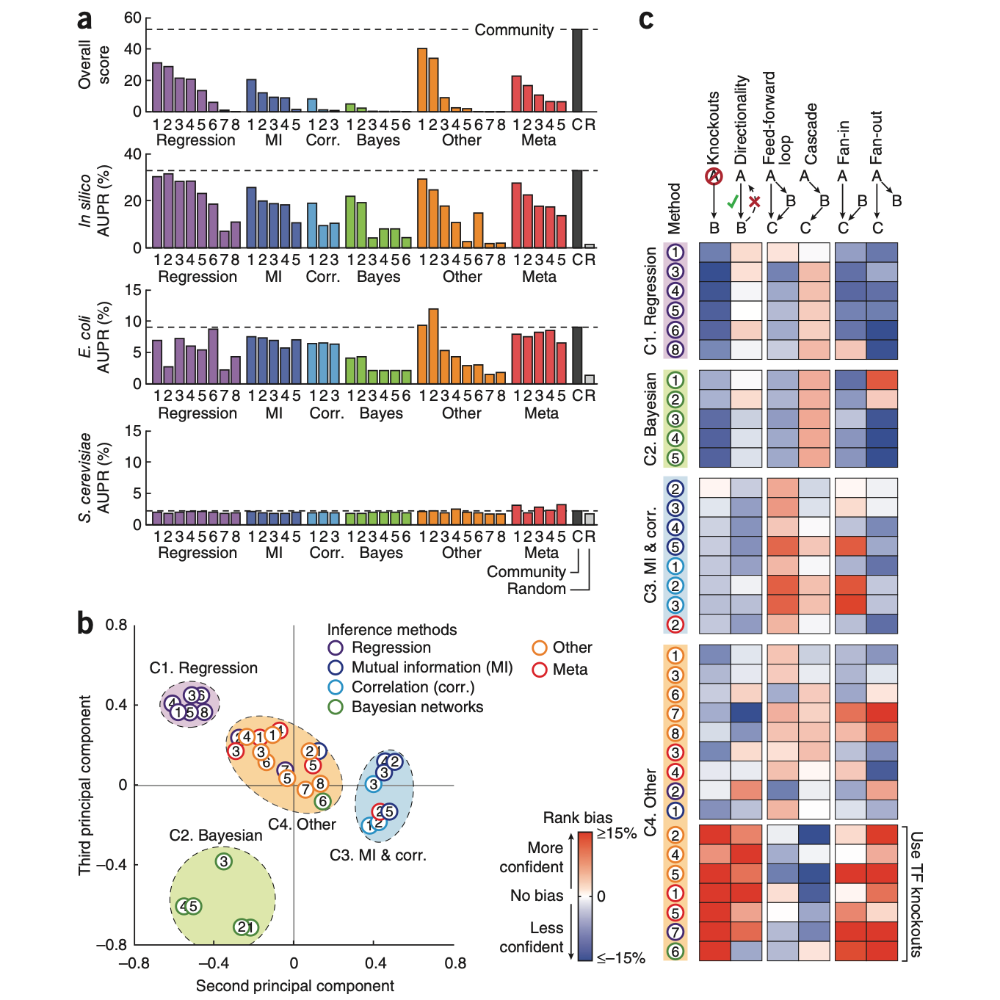
Wisdom of crowds for robust gene network inference. (pdf)
Journal: Nature Methods, 2012 Jul 15 doi: 10.1038/nmeth.2016 PMID:22796662 PMCID: PMC3512113 |
 |
71.
Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. (pdf)
Journal: Genome Research, 2012 Jul doi: 10.1101/gr.127191.111 PMID:22456606 PMCID: PMC3396374 |
 |
46.
SubMAP: aligning metabolic pathways with subnetwork mappings. (pdf)
Journal: Journal Of Computational Biology, 2011 Mar doi: 10.1089/cmb.2010.0280 PMID:21385030 PMCID: PMC3123932 |
 |
25.
The evolutionary dynamics of the Saccharomyces cerevisiae protein interaction network after duplication. (pdf)
Journal: Proceedings Of The National Academy Of Sciences Of The United States Of America, 2008 Jan 22 doi: 10.1073/pnas.0707293105 PMID:18199840 PMCID: PMC2242688 |
 |
18.
Reliable prediction of regulator targets using 12 Drosophila genomes. (pdf)
Journal: Genome Research, 2007 Dec doi: 10.1101/gr.7090407 PMID:17989251 PMCID: PMC2099599 |
Genome Evolution
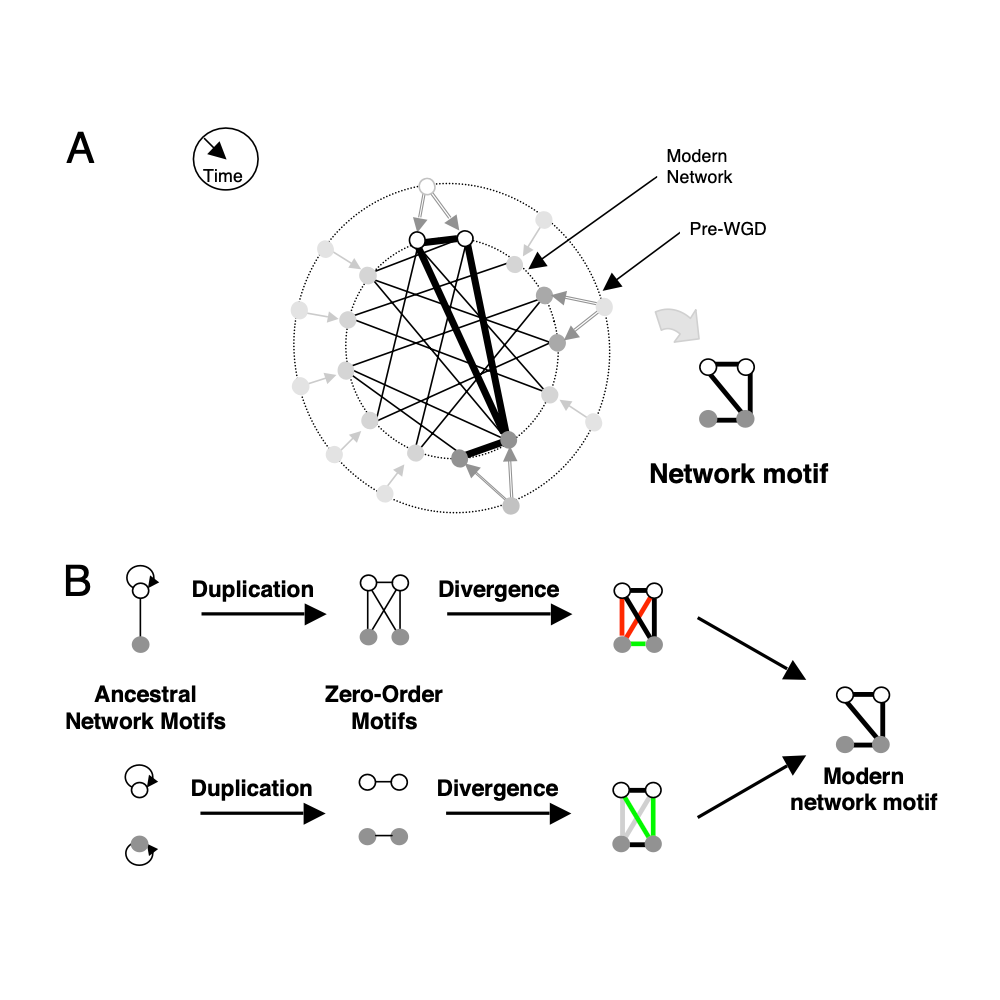
A unique and defining feature of biological systems is their ability to adapt to changing environments and evolve new functions and pathways, while constantly subjected to the forces of mutation and natural selection. Understanding the mechanisms associated with evolutionary innovation can teach us how to better interpret evolutionary signatures and comparative genomics evidence, yield new insights about the function of diverse genomic elements from their shared ancestry, and in the longer term reveal insights on how to engineer more evolvable and noise-tolerant systems. With the availability of many complete genomes, we have taken a new approach to studying gene evolution at the whole-genome level, which has resulted in numerous new insights that wouldn't be visible at the single-gene level. This revealed a unique genomic signature for whole-genome duplication which we demonstrated in yeast and fish, and has since been used to resolve the ancestry of many species, including our early vertebrate ancestors.
 |
267.
Genus-Wide Characterization of Bumblebee Genomes Provides Insights into Their Evolution and Variation in Ecological and Behavioral Traits. (pdf)
Journal: Molecular Biology And Evolution, 2021 Jan 23 doi: 10.1093/molbev/msaa240 PMID:32946576 PMCID: PMC7826183 |
 |
259.
SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. (pdf)
Journal: Research Square, 2020 Oct 1 doi: 10.21203/rs.3.rs-80345/v1 PMID:33024961 PMCID: PMC7536840 |
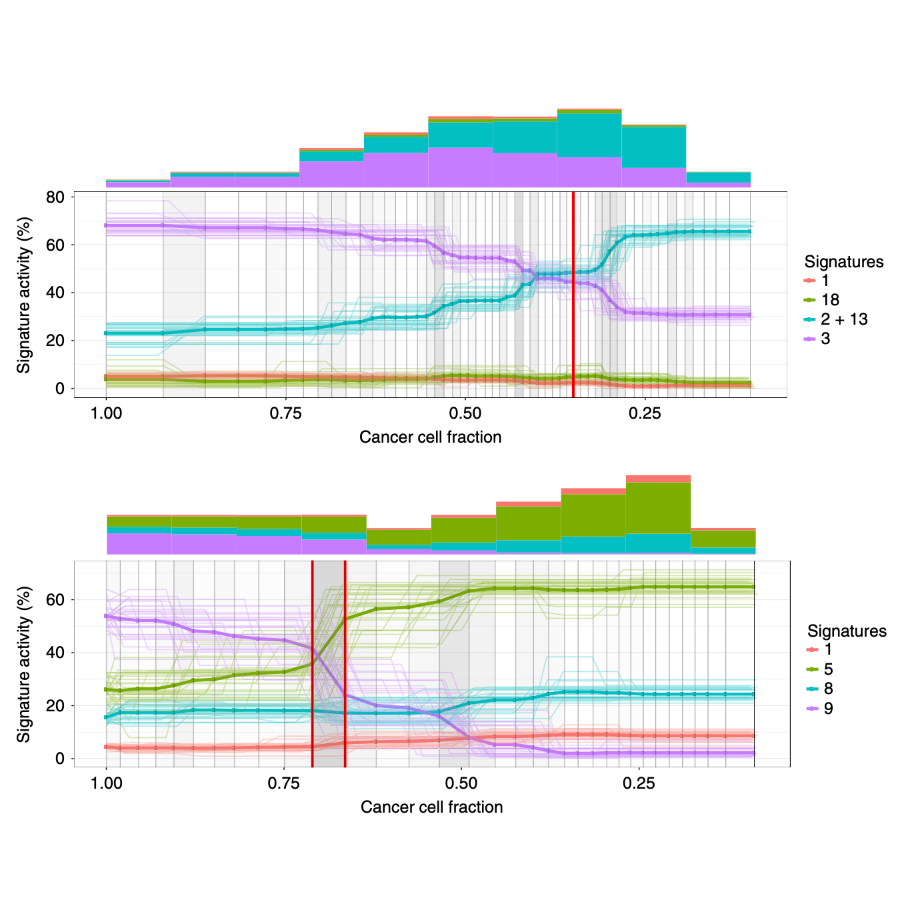 |
223.
Reconstructing evolutionary trajectories of mutation signature activities in cancer using TrackSig. (pdf)
Journal: Nature Communications, 2020 Feb 5 doi: 10.1038/s41467-020-14352-7 PMID:32024834 PMCID: PMC7002414 |
 |
218.
The evolutionary history of 2,658 cancers. (pdf)
Journal: Nature, 2020 Feb doi: 10.1038/s41586-019-1907-7 PMID:32025013 PMCID: PMC7054212 |
 |
129.
Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. (pdf)
Journal: Science, 2015 Jan 2 doi: 10.1126/science.1258522 PMID:25554792 PMCID: PMC4380271 |
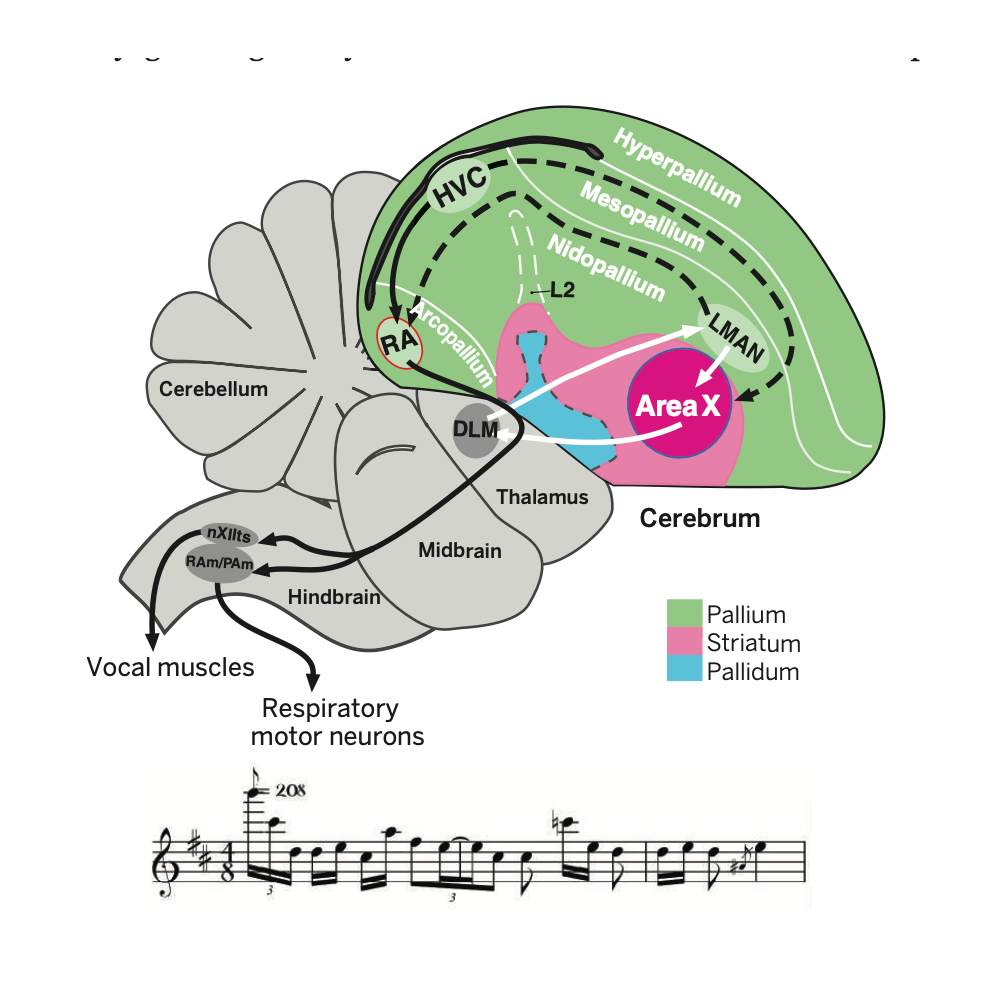 |
128.
Core and region-enriched networks of behaviorally regulated genes and the singing genome. (pdf)
Journal: Science, 2014 Dec 12 doi: 10.1126/science.1256780 PMID:25504732 PMCID: PMC4359888 |
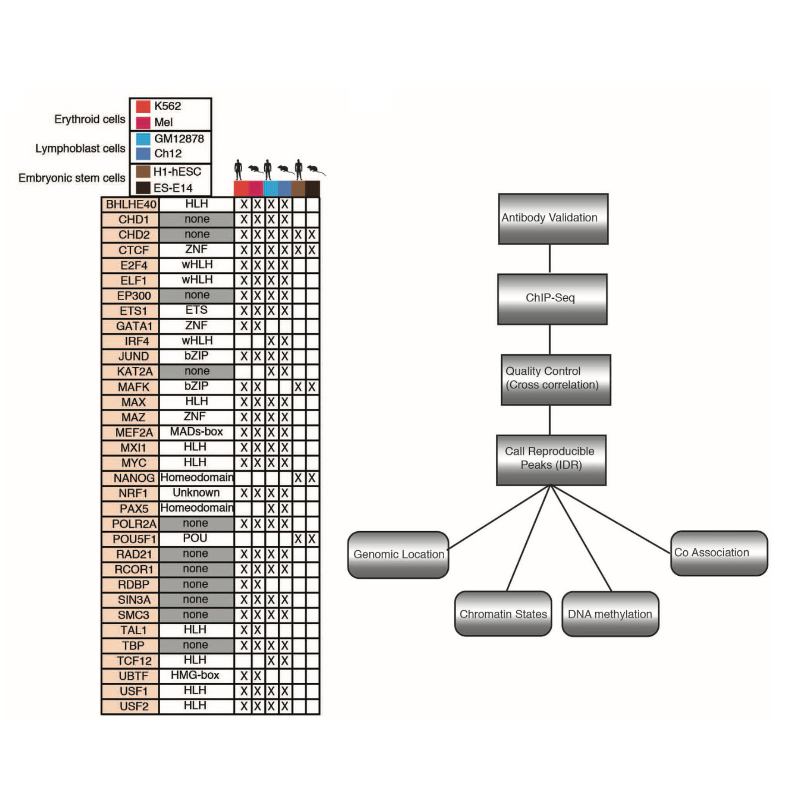 |
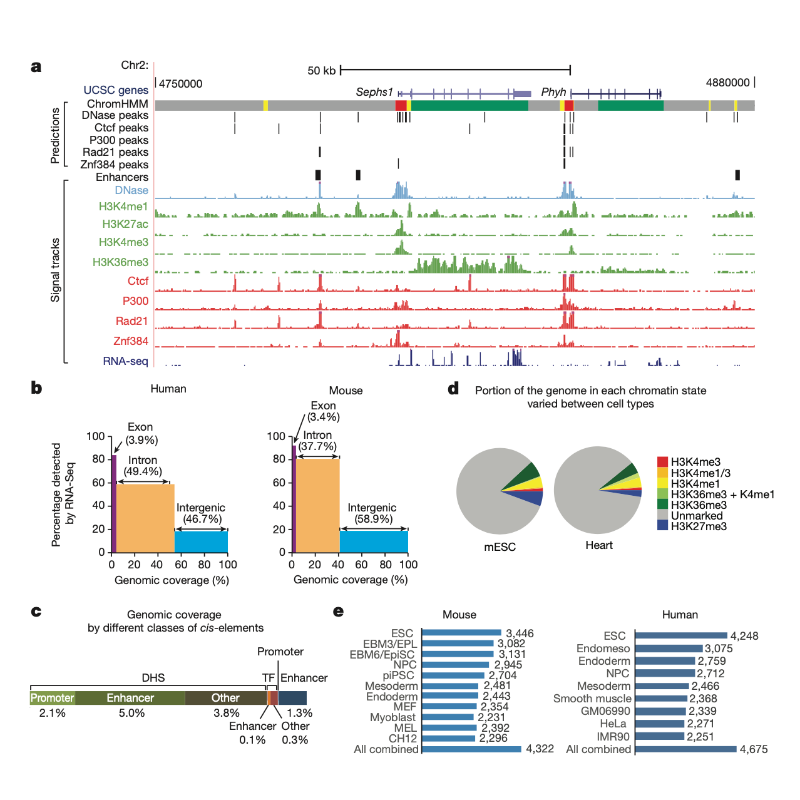
127.
Principles of regulatory information conservation between mouse and human. (pdf)
Journal: Nature, 2014 Nov 20 doi: 10.1038/nature13985 PMID:25409826 PMCID: PMC4343047 |
 |
126.
A comparative encyclopedia of DNA elements in the mouse genome. (pdf)
Journal: Nature, 2014 Nov 20 doi: 10.1038/nature13992 PMID:25409824 PMCID: PMC4266106 |
 |
99.
Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. (pdf)
Journal: Nature, 2013 Aug 22 doi: 10.1038/nature12326 PMID:23873043 |
 |
76.
High depth, whole-genome sequencing of cholera isolates from Haiti and the Dominican Republic. (pdf)
Journal: Bmc Genomics, 2012 Sep 11 doi: 10.1186/1471-2164-13-468 PMID:22963323 PMCID: PMC3473251 |
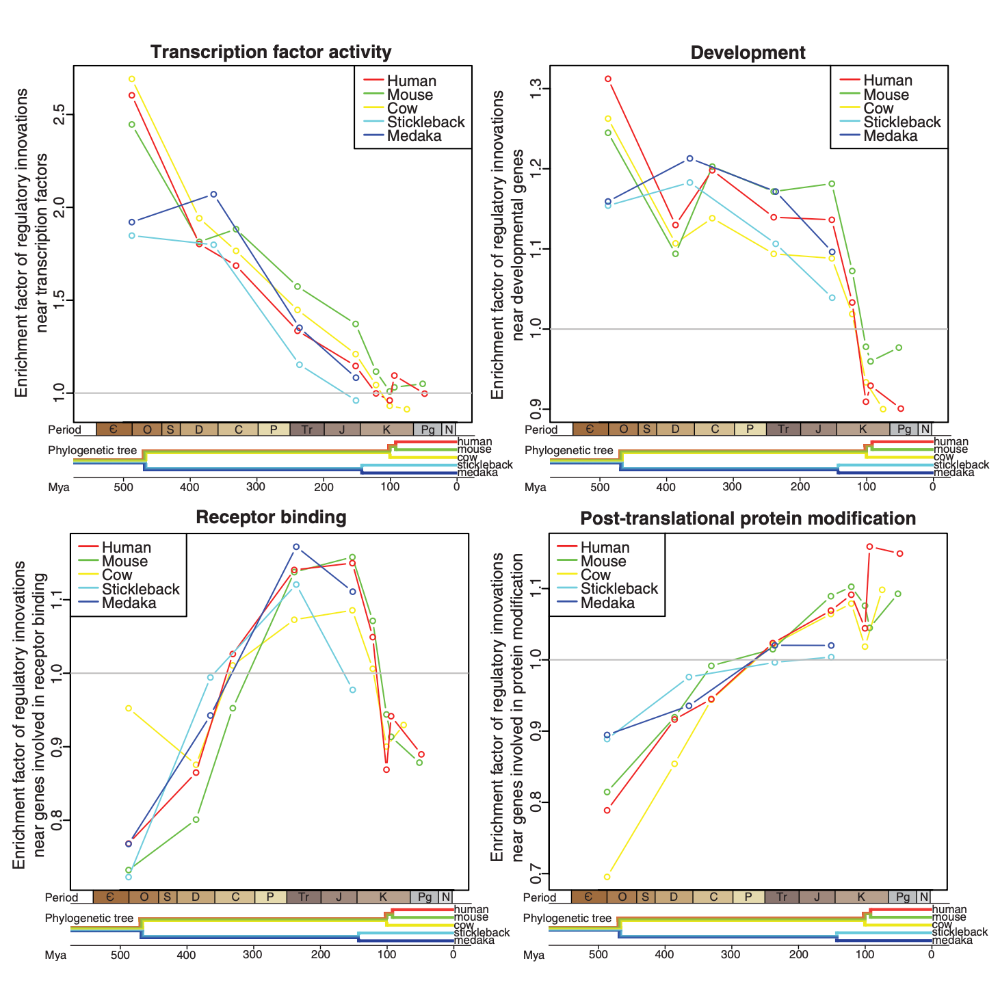 |
55.
Three periods of regulatory innovation during vertebrate evolution. (pdf)
Journal: Science, 2011 Aug 19 doi: 10.1126/science.1202702 PMID:21852499 PMCID: PMC3511857 |
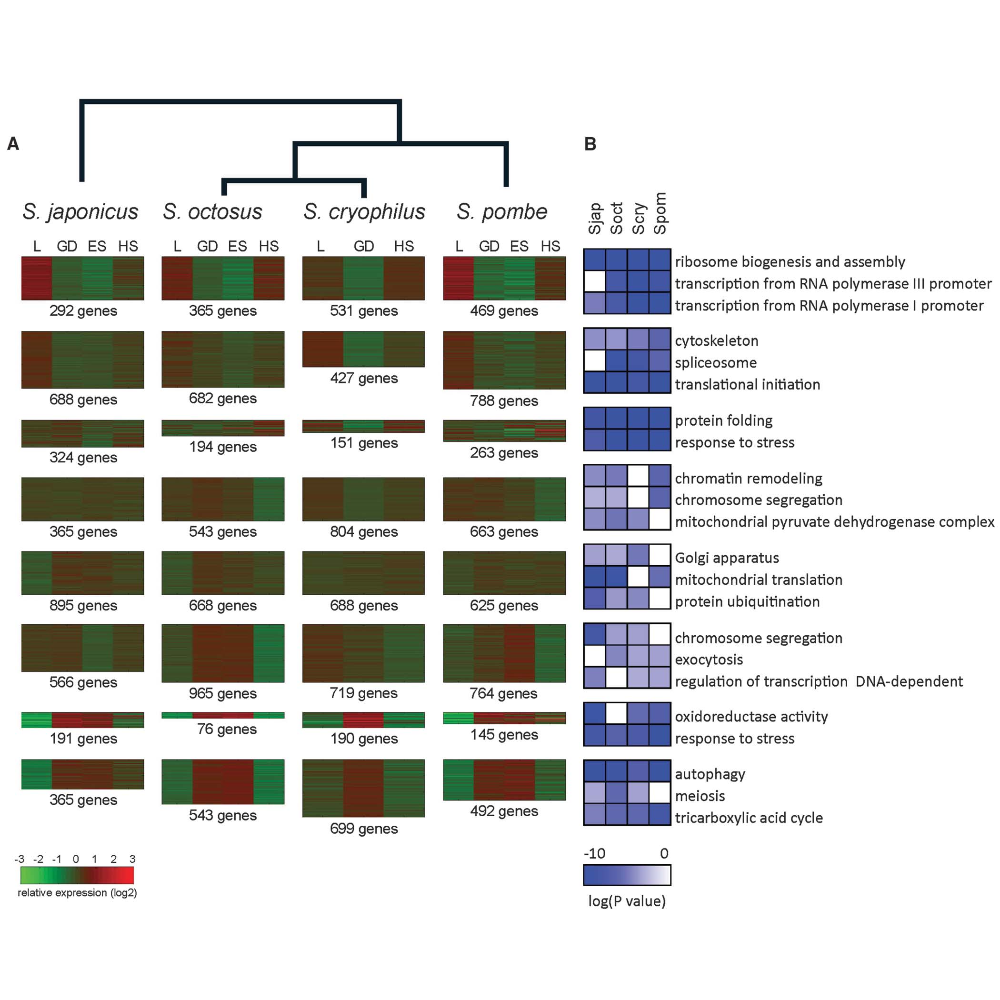 |
52.
Comparative functional genomics of the fission yeasts. (pdf)
Journal: Science, 2011 May 20 doi: 10.1126/science.1203357 PMID:21511999 PMCID: PMC3131103 |
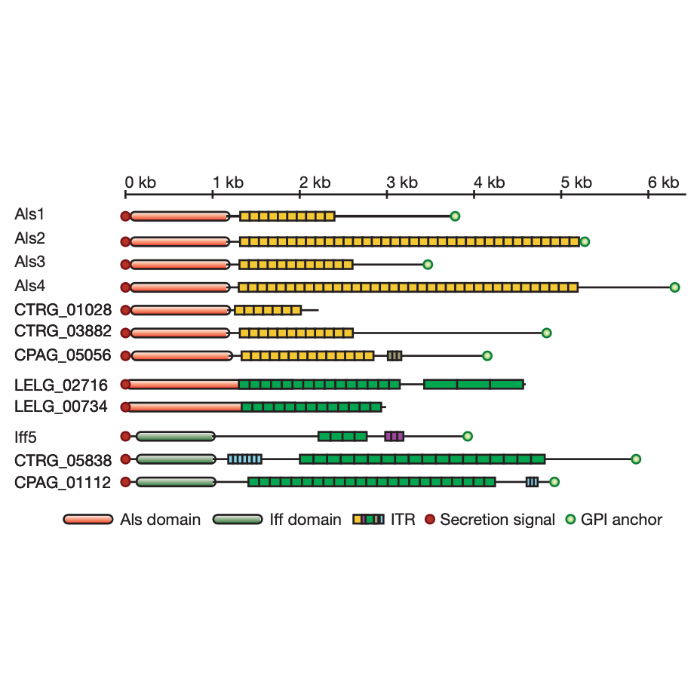 |
32.
Evolution of pathogenicity and sexual reproduction in eight Candida genomes. (pdf)
Journal: Nature, 2009 Jun 4 doi: 10.1038/nature08064 PMID:19465905 PMCID: PMC2834264 |
 |
27.
Genome analysis of the platypus reveals unique signatures of evolution. (pdf)
Journal: Nature, 2008 May 8 doi: 10.1038/nature06936 PMID:18464734 PMCID: PMC2803040 |
 |
16.
Evolution of genes and genomes on the Drosophila phylogeny. (pdf)
Journal: Nature, 2007 Nov 8 doi: 10.1038/nature06341 PMID:17994087 |
 |
8.
Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. (pdf)
Journal: Nature, 2004 Oct 21 doi: 10.1038/nature03025 PMID:15496914 |
 |
6.
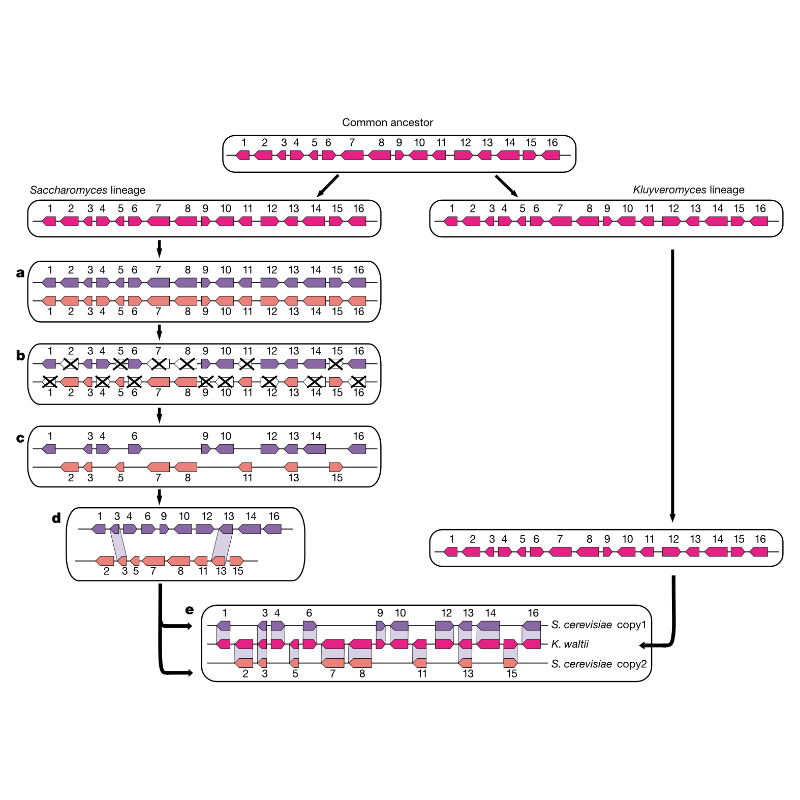
Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. (pdf)
Journal: Nature, 2004 Apr 8 doi: 10.1038/nature02424 PMID:15004568 |
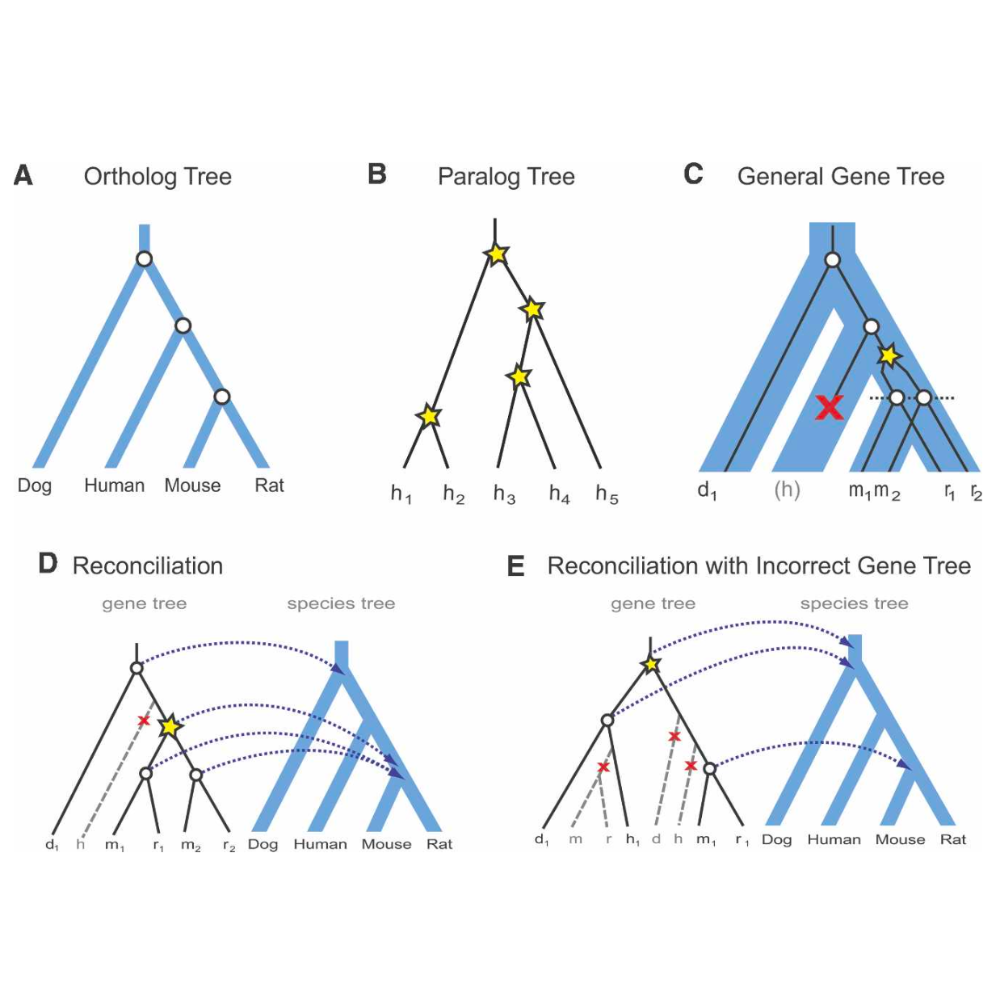
Phylogenomics
We have used the availability of many closely-related genomes to go beyond traditional phylogenetic methods and develop new 'phylogenomic' methods for resolving gene phylogenies across complete genomes jointly, enabling much more accurate inference methods, by recognizing two distinct evolutionary forces, one acting at the gene level, and one at the genome level. We applied these methods to Candida genomes, including both strong and weak human pathogens, and revealed new candidate virulence genes that are rapidly evolving and expanding in pathogens, thus expanding potential drug target genes.
 |
190.
RANGER-DTL 2.0: rigorous reconstruction of gene-family evolution by duplication, transfer and loss. (pdf)
Journal: Bioinformatics, 2018 Sep 15 doi: 10.1093/bioinformatics/bty314 PMID:29688310 PMCID: PMC6137995 |
 |
139.
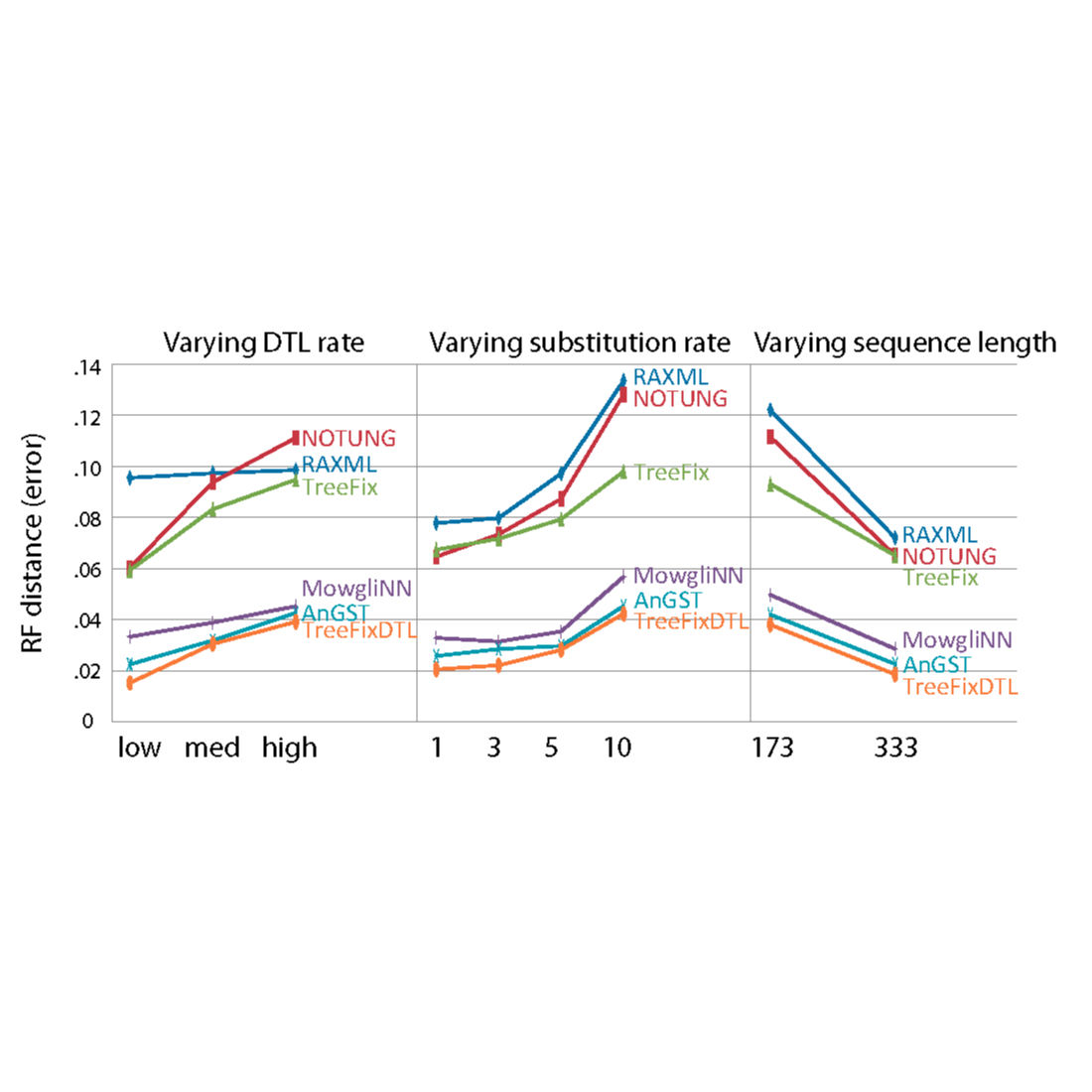
Improved gene tree error correction in the presence of horizontal gene transfer. (pdf)
Journal: Bioinformatics, 2015 Apr 15 doi: 10.1093/bioinformatics/btu806 PMID:25481006 PMCID: PMC4393519 |
 |
114.
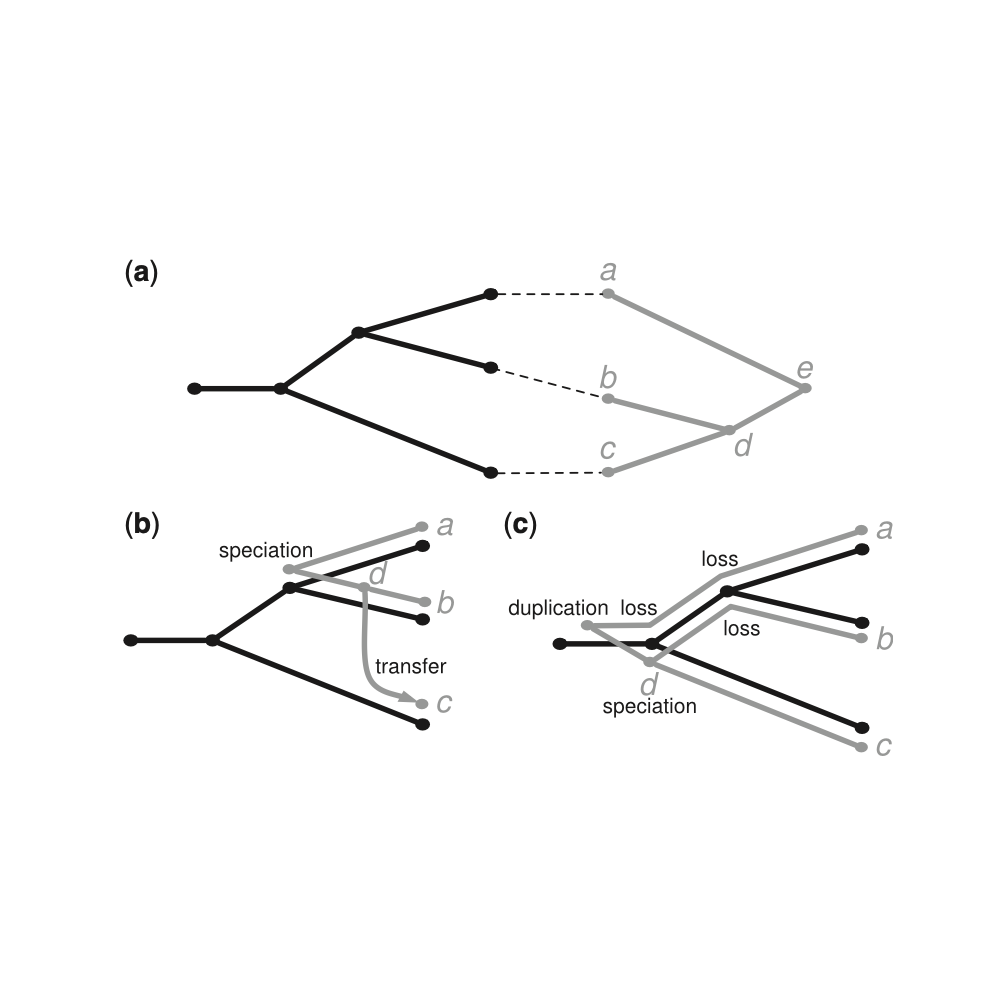
Pareto-optimal phylogenetic tree reconciliation. (pdf)
Journal: Bioinformatics, 2014 Jun 15 doi: 10.1093/bioinformatics/btu289 PMID:24932009 PMCID: PMC4058917 |
 |
107.
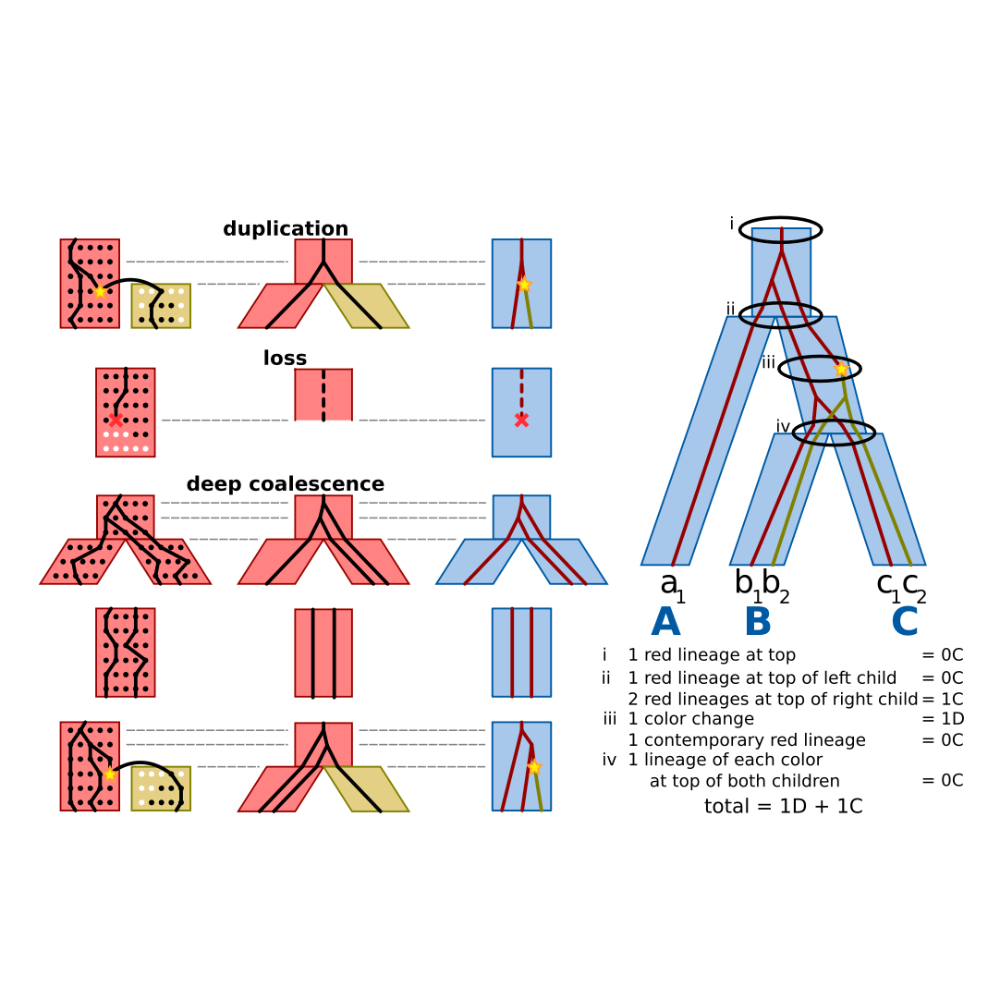
Most parsimonious reconciliation in the presence of gene duplication, loss, and deep coalescence using labeled coalescent trees. (pdf)
Journal: Genome Research, 2014 Mar doi: 10.1101/gr.161968.113 PMID:24310000 PMCID: PMC3941112 |
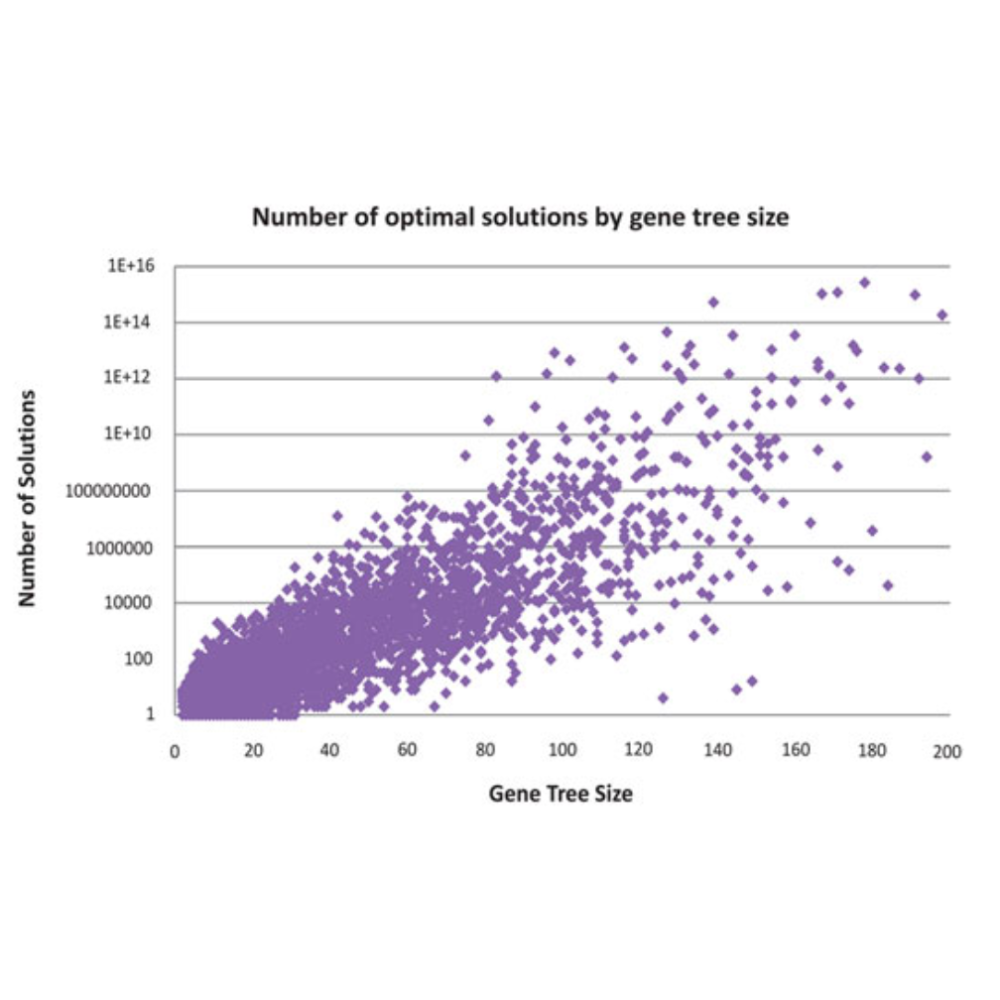 |
100.
Reconciliation revisited: handling multiple optima when reconciling with duplication, transfer, and loss. (pdf)
Journal: Journal Of Computational Biology, 2013 Oct doi: 10.1089/cmb.2013.0073 PMID:24033262 PMCID: PMC3791060 |
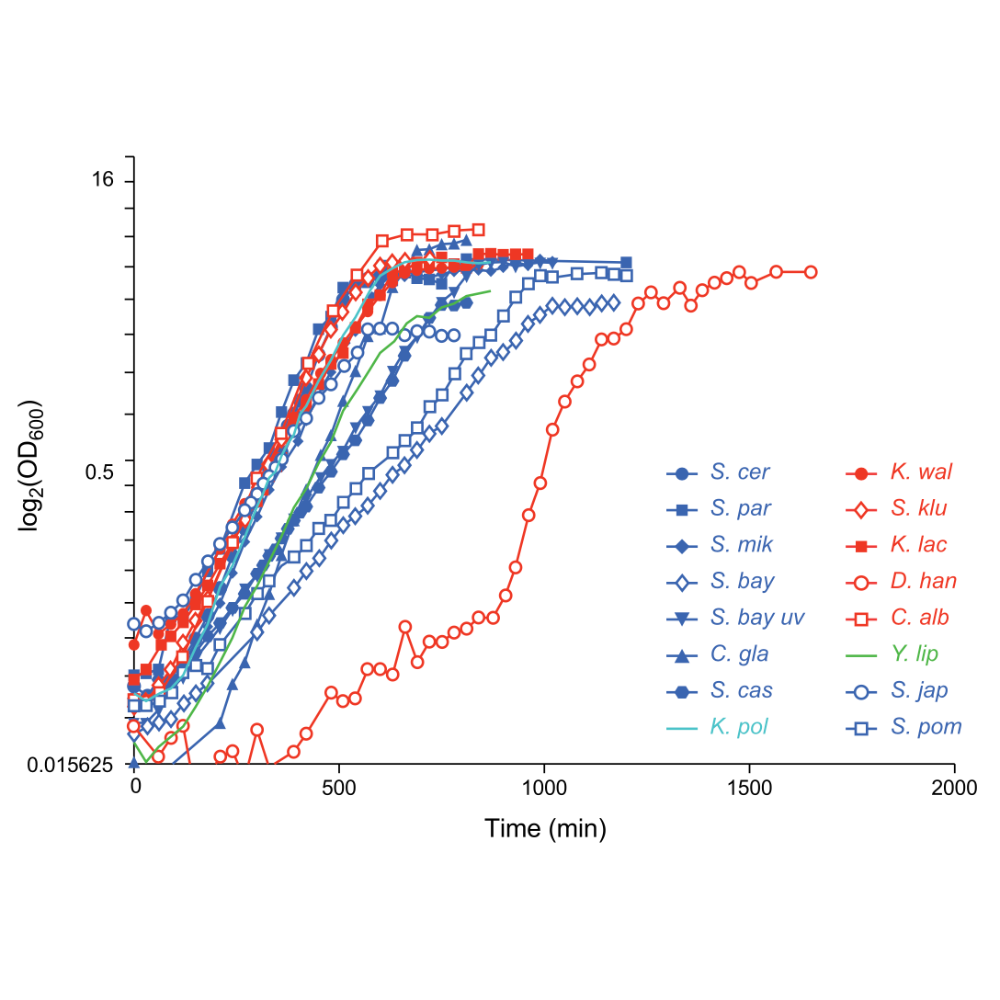 |
95.
Evolutionary principles of modular gene regulation in yeasts. (pdf)
Journal: Elife, 2013 Jun 18 doi: 10.7554/eLife.00603 PMID:23795289 PMCID: PMC3687341 |
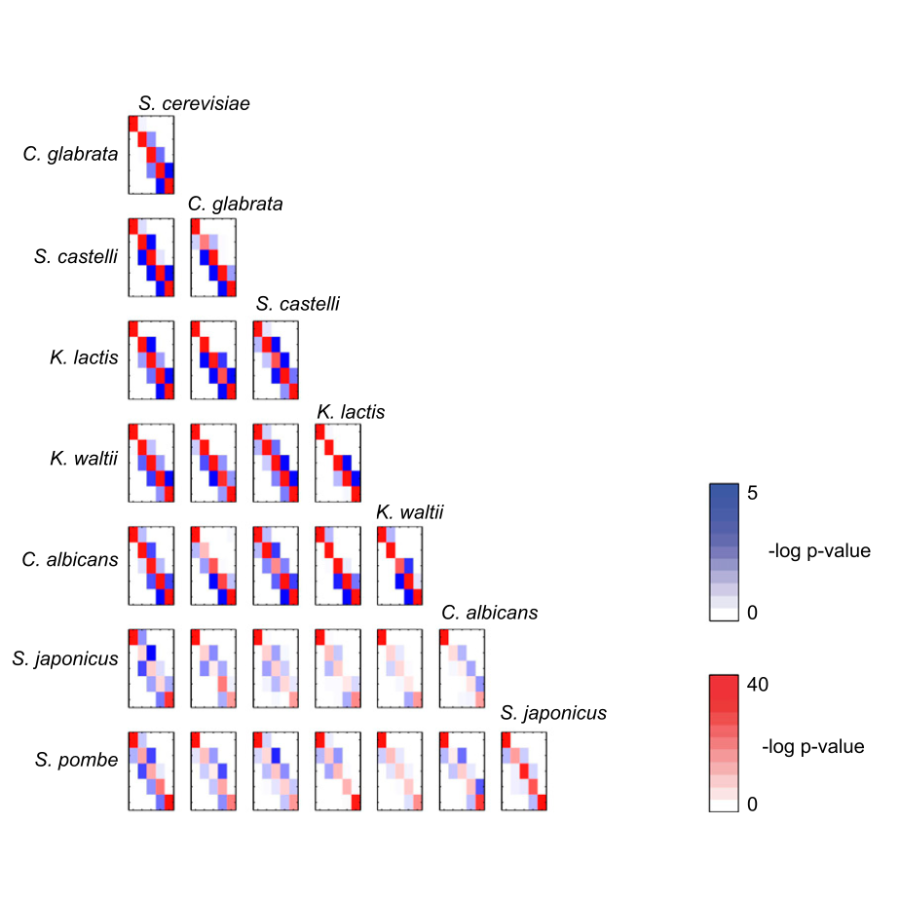 |
93.
Arboretum: reconstruction and analysis of the evolutionary history of condition-specific transcriptional modules. (pdf)
Journal: Genome Research, 2013 Jun doi: 10.1101/gr.146233.112 PMID:23640720 PMCID: PMC3668358 |
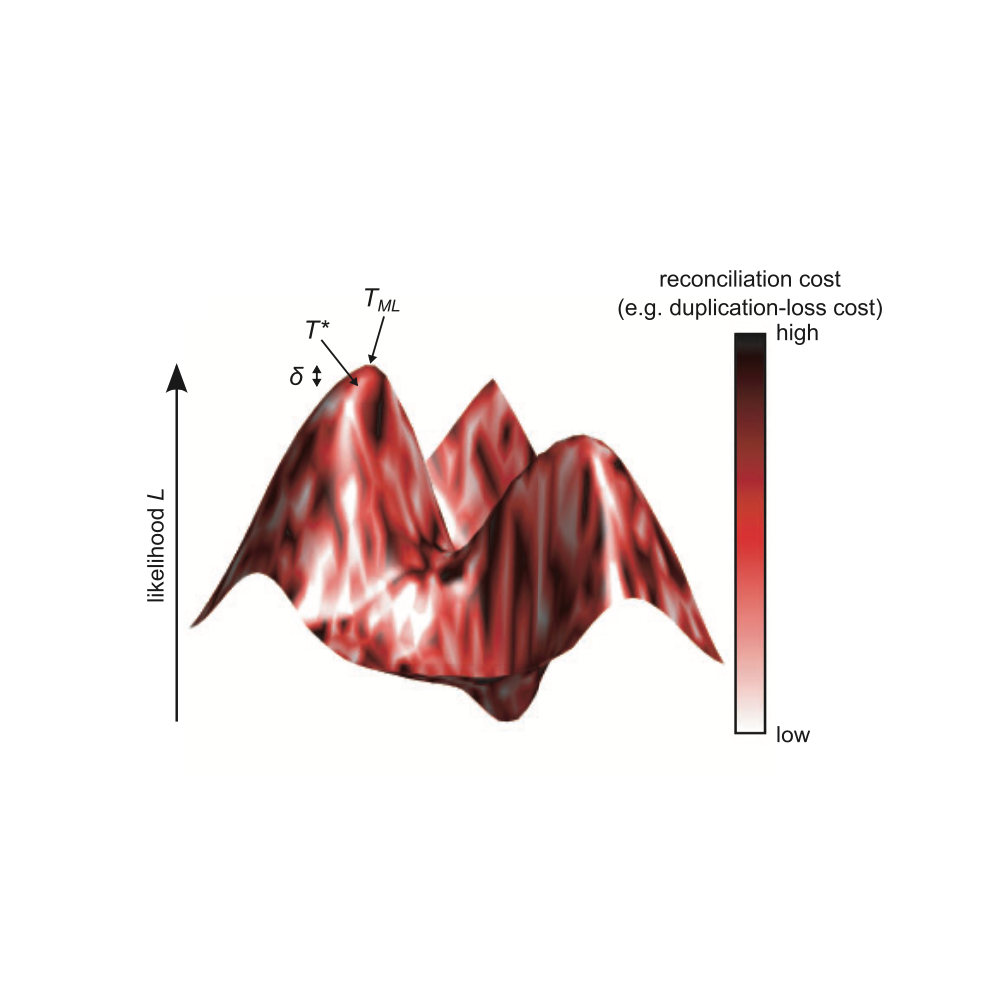 |
84.
TreeFix: statistically informed gene tree error correction using species trees. (pdf)
Journal: Systematic Biology, 2013 Jan 1 doi: 10.1093/sysbio/sys076 PMID:22949484 PMCID: PMC3526801 |
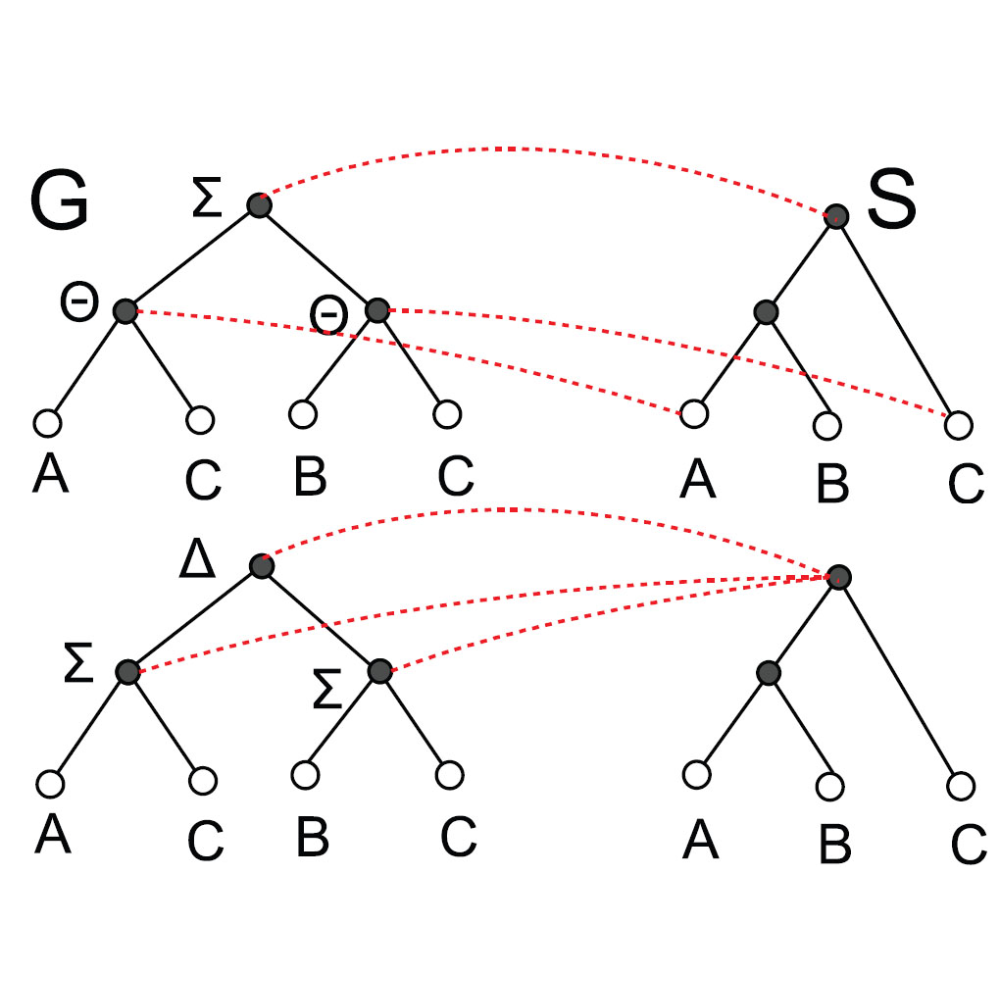 |
70.
Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss. (pdf)
Journal: Bioinformatics, 2012 Jun 15 doi: 10.1093/bioinformatics/bts225 PMID:22689773 PMCID: PMC3371857 |
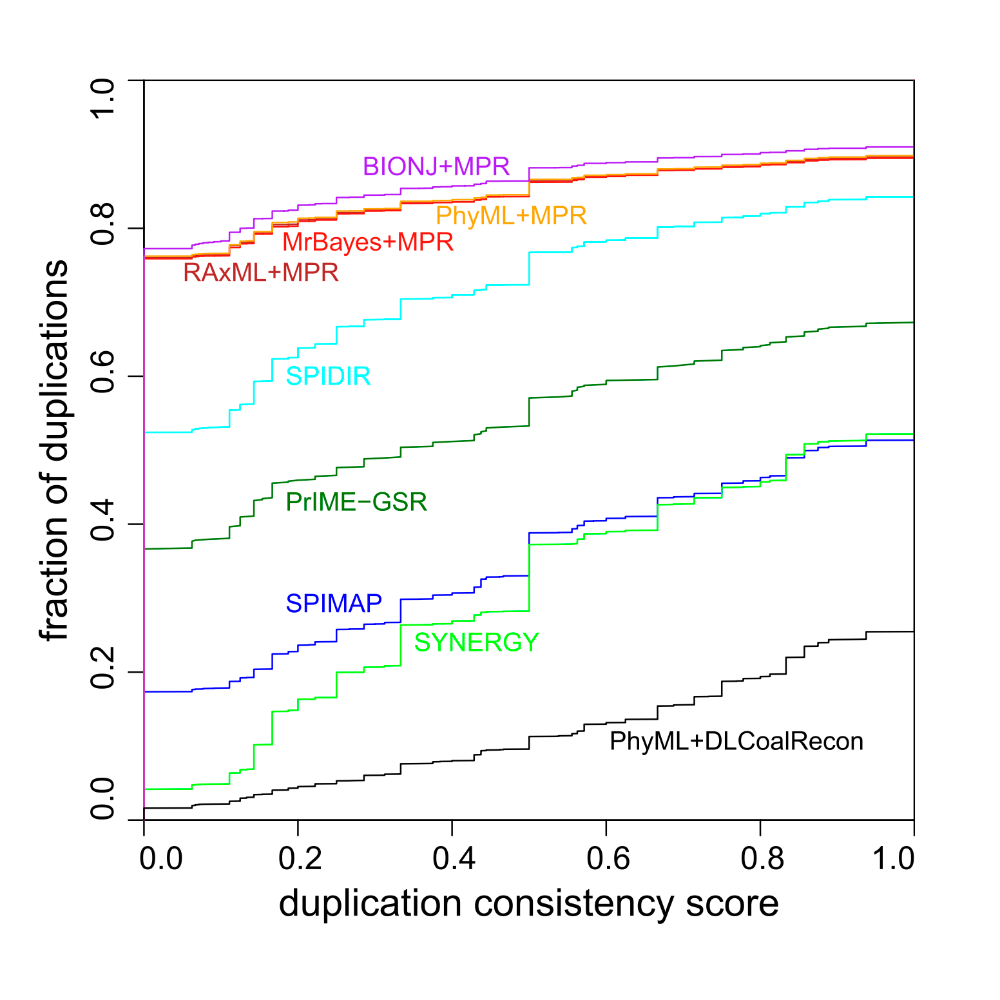 |
68.
Unified modeling of gene duplication, loss, and coalescence using a locus tree. (pdf)
Journal: Genome Research, 2012 Apr doi: 10.1101/gr.123901.111 PMID:22271778 PMCID: PMC3317157 |
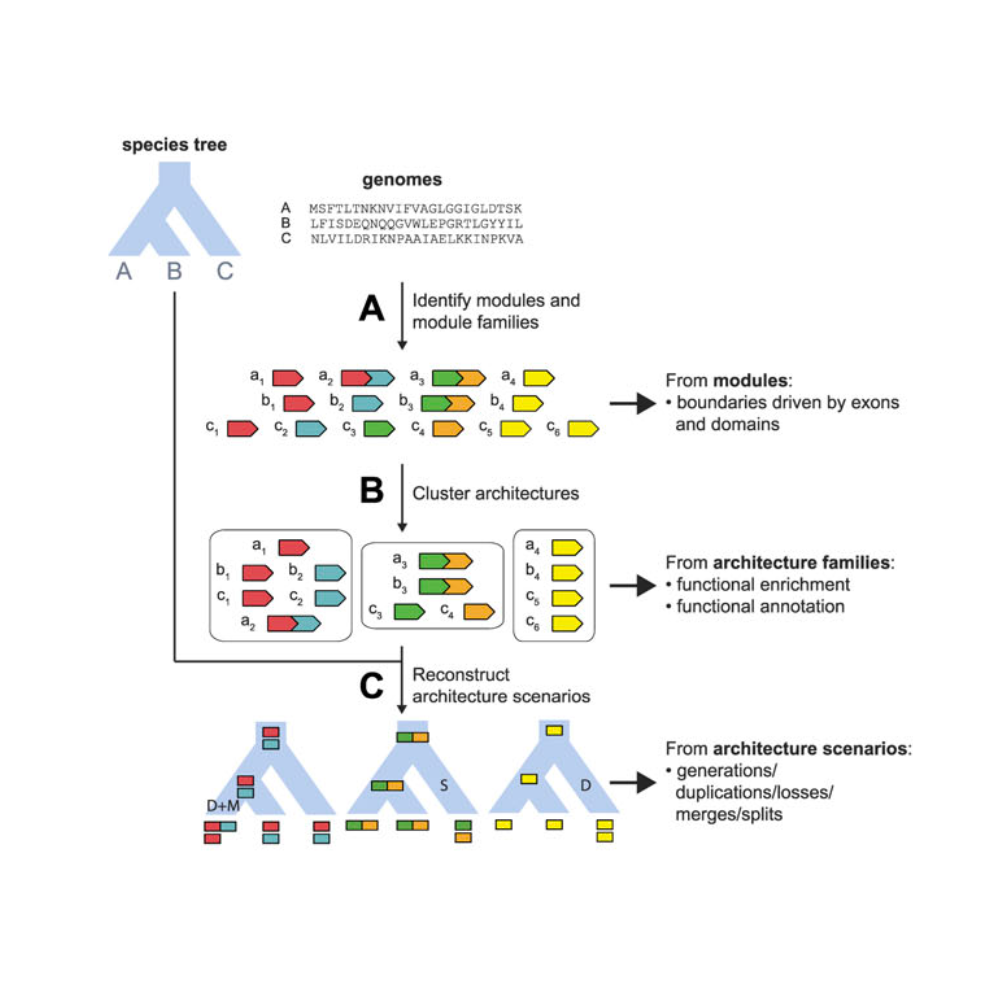 |
65.
Evolution at the subgene level: domain rearrangements in the Drosophila phylogeny. (pdf)
Journal: Molecular Biology And Evolution, 2012 Feb doi: 10.1093/molbev/msr222 PMID:21900599 PMCID: PMC3258039 |
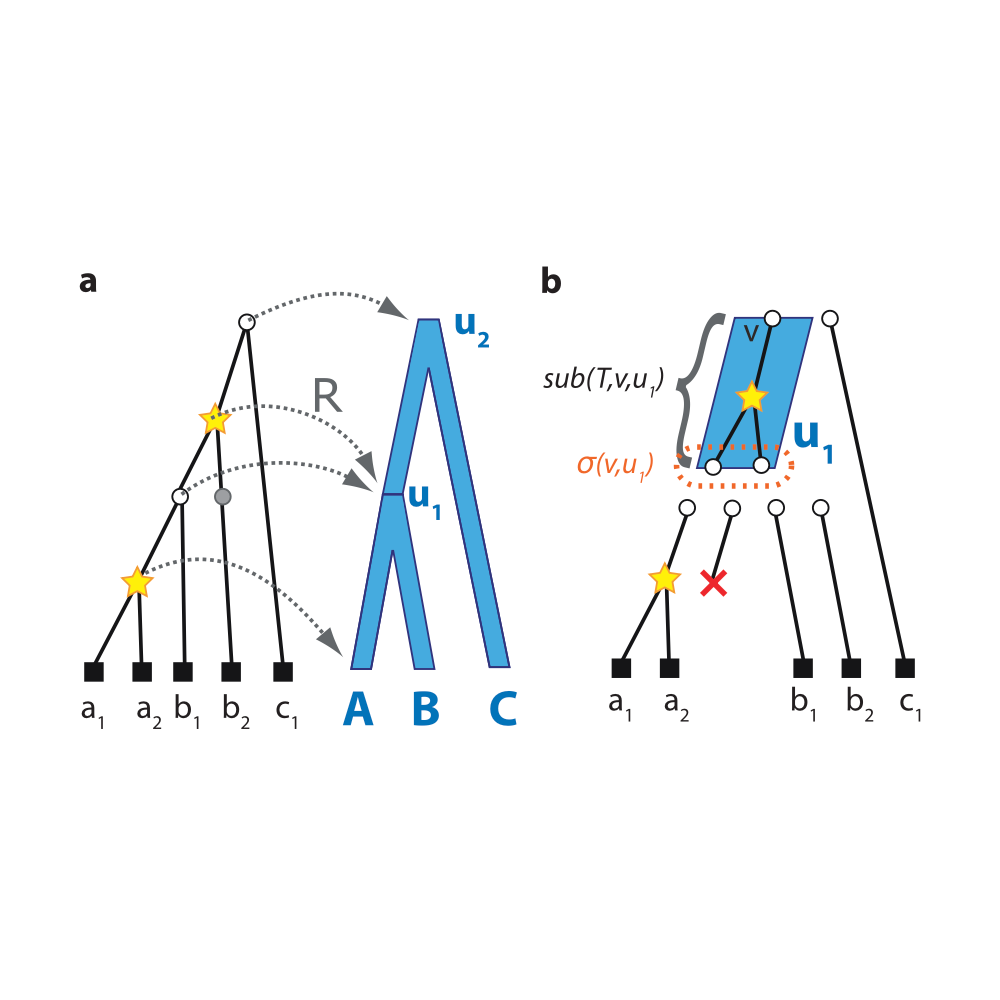 |
43.
A Bayesian approach for fast and accurate gene tree reconstruction. (pdf)
Journal: Molecular Biology And Evolution, 2011 Jan doi: 10.1093/molbev/msq189 PMID:20660489 PMCID: PMC3002250 |
 |
22.
Accurate gene-tree reconstruction by learning gene- and species-specific substitution rates across multiple complete genomes. (pdf)
Journal: Genome Research, 2007 Dec doi: 10.1101/gr.7105007 PMID:17989260 PMCID: PMC2099600 |
Algorithmic Papers in Computation Biology
While most of our papers have been in biological journals and in collaboration with experimental investigators, we have also written a small number of computational papers, for computational biology conferences or computational journals. These have frequently introduced new algorithms that we have independently or in parallel applied to biological datasets, and other times provided more details on the methods underlying some of our genomics results. These papers will be of particular interest to computational scientists seeking to gain a deeper understanding of the computational challenges associated with the genomics problems we are working on. The algorithms presented in these papers include methods for ortholog and paralog determination, gene identification from their evolutionary signatures, motif discovery and clustering, network motif discovery using symmetry breaking conditions, and network inference methods.
 |
67.
ChromHMM: automating chromatin-state discovery and characterization. (pdf)
Journal: Nature Methods, 2012 Feb 28 doi: 10.1038/nmeth.1906 PMID:22373907 PMCID: PMC3577932 |
 |
54.
PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. (pdf)
Journal: Bioinformatics, 2011 Jul 1 doi: 10.1093/bioinformatics/btr209 PMID:21685081 PMCID: PMC3117341 |
 |
46.
SubMAP: aligning metabolic pathways with subnetwork mappings. (pdf)
Journal: Journal Of Computational Biology, 2011 Mar doi: 10.1089/cmb.2010.0280 PMID:21385030 PMCID: PMC3123932 |
 |
45.
Error and error mitigation in low-coverage genome assemblies. (pdf)
Journal: Plos One, 2011 Feb 14 doi: 10.1371/journal.pone.0017034 PMID:21340033 PMCID: PMC3038916 |
 |
5.
Methods in comparative genomics: genome correspondence, gene identification and regulatory motif discovery. (pdf)
Journal: Journal Of Computational Biology, 2004 doi: 10.1089/1066527041410319 PMID:15285895 |
 |
1.
Phylogenetically and spatially conserved word pairs associated with gene-expression changes in yeasts. (pdf)
Journal: Genome Biology, 2003 doi: 10.1186/gb-2003-4-7-r43 PMID:12844359 PMCID: PMC193630 |
Computational Geometry / Vision / Graphics
Prior to computational biology, Prof. Kellis worked in several areas of computer science, and in particular computational geometry, graphics, and vision. He co-developed an algorithm for surface reconstruction (which has received 664 citations), methods for polygon and curve morphing, shape-recognition methods for sketch-based image retrieval, and efficient representations for online handwritten character recognition.
Other Manuscripts (Robotics / AI)
Other areas of interest included robotics and artificial intelligence, resulting in projects on robot programming, developing an interactive robot programming framework for kids, pattern classification methods for music or the stock market based on attentional states, and other past research.
Editorials
Lastly, Prof Kellis was recruited a few times to editors to write reviews or meeting reports.
 |
35.
Sequences to systems. (pdf)
Journal: Genome Biology, 2010 doi: 10.1186/gb-2010-11-5-303 PMID:20500907 PMCID: PMC2898084 |
 |
4.
The changing face of genomics. (pdf)
Journal: Genome Biology, 2004 doi: 10.1186/gb-2004-5-5-324 PMID:15128443 PMCID: PMC416465 |