Alzheimer’s disease multi-region data repository
Overview:
This is a supplementary website for: Single-cell multiregion dissection of Alzheimer’s disease
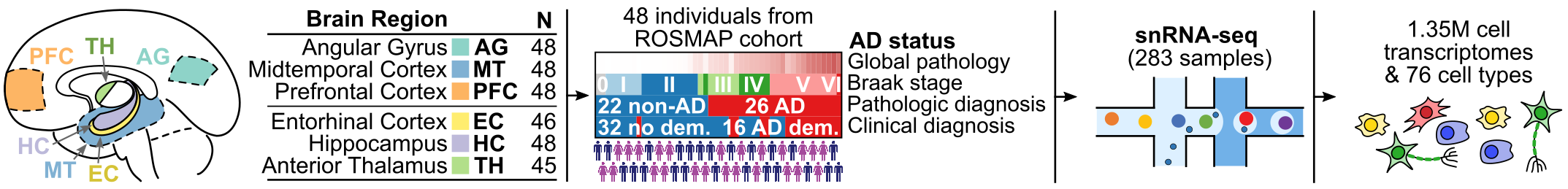
We profiled and annotated a multi-region single-cell transcriptomic atlas of the aged human brain covering 1.3 million cells from 283 post-mortem human brain samples across six brain regions, 48 individuals, and six stages of AD progression to study molecular mechanisms of AD progression across brain regions.
- In addition to resources provided below and on Synapse, the single-cell transcriptomic dataset is available for interactive visualization through the UCSC Cell Browser.
- Summary analyses are currently available as supplementary tables below.
- We also make results including expression profiles, AD differential changes, and gene modules available interactively through compbio2.mit.edu/ad_multiregion/.
- Analysis code for the paper can be found on github or on zenodo.
Contact: Hansruedi Mathys - mathysh at pitt dot edu / Carles Boix - cboix at mit dot edu / Leyla Akay - leyla at mit dot edu
Modules (scdemon)
The paper also presents a method, scdemon (single-cell decorrelated module networks), for discovering gene modules from large-scale single-cell expression data.
You can find the python package at https://github.com/KellisLab/scdemon, and install it from github as:
pip install git+https://github.com/KellisLab/scdemon
More detailed instructions and documentation can be found at https://scdemon.readthedocs.io/ or on the README at github.
Module definitions for the single-cell Alzheimer’s multi-region atlas can be found here.
Datasets
Complete human raw data are released in collaboration with ROSMAP through Synapse (syn52293442). The data and human metadata must be accessed through Synapse with a Data Use Certificate (DUC). Human metadata can be found at Synapse (syn3157322) or can be requested through the RADC Hub.
Datasets corresponding to manuscript analyses can be found here and are summarized in the table below. Processed human datasets have been deidentified to protect confidentiality - the mapping to ROSMAP IDs and complete metadata can be found on Synapse.
| Dataset | Files | Description |
|---|---|---|
| Sample metadata | Table in tsv | Sample metadata table (Supplementary Table 1) |
| Markers | Table in xlsx | Gene expression marker lists (Supplementary Table 2) |
| cNMF programs | Table in xlsx | Gene expression programs from cNMF (Supplementary Table 3) |
| SCENIC regulons | Table in xlsx | Transcriptional regulons from SCENIC (Supplementary Table 4) |
| Neuron scores | Table in xlsx | Scores for GABAergic, glutamatergic, and projecting neurons (Supp. Table 5) |
| Modules (scdemon) | Assignments: (xlsx, Rds), enrichments (xlsx, tsv) and directory with additional info | Gene expression module assignments from scdemon (Supplementary Table 6), module covariate enrichments (Supplementary Table 7), and pseudobulk datasets, enrichments for AD DEGs, coordinates, and other information (in directory). |
| Markers for vulnerability | Table in xlsx | Genes associated with subtype resilience and vulnerability for both excitatory and inhibitory neurons (Supplementary Table 8) |
| Differentially expressed genes | Tarball of tables tar.gz and directory with tables | Differential gene expression tables (Supp. Table 9) for AD pathology or AD diagnosis, aggregated by cell type. Also includes a table for sex*AD interaction. |
| Pathology-biased DEGs | Table in tsv | Pathology-biased differentially expressed genes, for neuritic plaque or NFT burden (Supplementary Table 10) |
| Cognitive Resilience DEGs | Tables in directory | Differentially expressed genes for measures of cognitive resilience (CR) and cognitive decline resilience (CDR) in each cell type, against measures of pathology and AD diagnosis |
Other studies
Multiple studies from our group have incorporated this resource into their investigation of different aspects of Alzheimer’s disease and may be of interest:
- Microglial state dynamics in AD (Sun, Victor, et al. 2023) // (website)
- Epigenomic landscape and its erosion in AD (Xiong, James, Boix et al., 2023) // (website)
- AD and aging brain (Mathys, Peng, Boix, et al. 2023) // (website)
- Impact of neuronal DSBs on neurodegeneration (Dileep, Boix, et al. 2023) // (website)
- Transcriptomic dissection of AD vasculature (Sun et al. 2023) // (website)