The SPIMAP software package contains several programs reconstructing gene trees and inferring their gene duplications and losses. The concepts and algorithms behind the main program, spimap, are presented in our SPIMAP paper. This documentation describes how to use each of the programs in the software package and defines the various file formats that are used.
For an example of how to run these programs see the examples directory (e.g. examples/analyze-fungi.sh).

Note: This documentation is currently under development and will be regularly updated with more information about the SPIMAP software package.
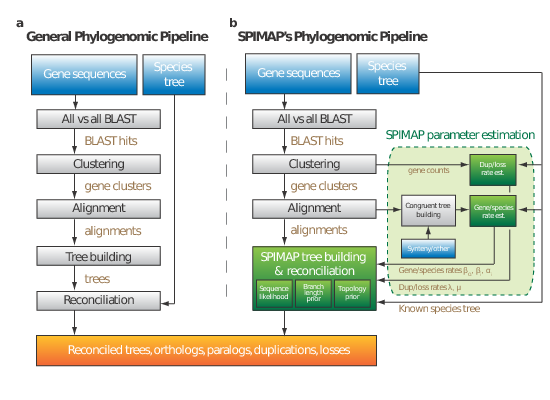
The main SPIMAP algorithm is implemented in the spimap program. The purpose of this program is reconstruct gene trees from DNA alignments. Accurately reconstructing gene trees is a challenging problem. SPIMAP's strategy is to more accurately reconstruct gene trees by using additional information learned from the species tree and the genome.
This genome-wide information is captured in two sets of parameters that the spimap program needs. The first set are gene duplication and loss rates (λ and μ). You can either specify these rates based on previously performed studies, or you can use the spimap-train-duploss program to learn them from gene counts.
The second set of parameters needed by spimap are substitution rate parameters (βG, α, β). These are estimated from a small subset of trusted purely orthologous gene trees. Once these parameters are learned, they can be used to aid in the reconstruction of more difficult paralogous families. Tthe spimap-train-rates program can be used to estimate these parameters.
The figure below illustrates a computational pipeline for how the three programs spimap, spimap-train-duploss, and spimap-train-rates work together. Additionally, this package also includes several other helper programs that can prepare data sets for training (see spimap-prep-duploss and spimap-prep-rates).
This specifies the DNA alignment to use for reconstructing a gene tree. The file should be in FASTA format.
This specifies which species each gene belongs using the file species map written in in *.smap format.
This specifies a known species tree with the file species tree written in newick format.
This specifies the SPIMAP substitution rate parameters (βG, α, β) written in the *.param file format. This file is generated by spimap-train-rates. Optionally, this argument can be skipped, in which case a gene- and species-specific rates model will not be used to reconstruct the gene tree.
This specifies a prefix for all of the output files. By default the prefix is "spimap" (e.g. "spimap.tree", "spimap.recon", "spimap.log"). The prefix can also include a different output directory.
With this argument SPIMAP will output the reconciliation found to the file "<output filename prefix>.recon" written in the *.recon format.
This specifies the transition/transversion ratio κ for the HKY sequence model. By default κ will be estimated from the alignment.
This specifies the background base frequency for the HKY sequence model. By default these frequencies are estimated from the alignment.
This specifies the gene duplication rate. Commonly, this should be specified in units of duplications/gene/million years. See species tree section for a discussion of the unit of time. See spimap-train-duploss for a way to estimate this parameter from data.
This specifies the gene loss rate. Commonly, this should be specified in units of loss/gene/million years. See species tree section for a discussion of the unit of time. See spimap-train-duploss for a way to estimate this parameter from data.
This specifies the number of iterations SPIMAP should search for the gene tree.
This specifies the number of subproposals (default=50) for each main search iteration. Choosing a number between 100-1000 usually increases search efficiency, therefore allowing one to use fewer main iterations ("-i").
Use this argument to perform bootstrapping. The alignment will be resampled with replacement number bootstrap times and a gene tree will be reconstructed for each sample.
You can adjust the amount of logging/debugging information that SPIMAP displays by adjusting this argument (0=quiet, 1=low, 2=medium, 3=high)
This specifies a different file for saving log information. Use "-" to display on stdout (standard output).
When given, SPIMAP will display version information.
When given, SPIMAP will display help information.
This program prepares a gene count matrix file for learning gene duplication-loss rates. It can produce this file using one of two methods:
This specifies a known species tree with the file species tree written in newick format.
This specifies which species each gene belongs using the file species map written in in *.smap format.
This specifies the output filename for the total gene counts per species. The format is a tab delimited matrix, where each row represents a gene family and each column represents a species. Each cell indicates the number of genes in that family and species.
If input method 2 is used (see above), this specifies a gene partition file. The format is a tab delimited. Each line represents a single family, and the line should contain each gene that appears in that family. Genes can be listed in any order.
When given, shows this help message.
This program learns gene duplication-loss rates from a gene counts matrix file.
This specifies a known species tree with the file species tree written in newick format.
This specifies which species each gene belongs using the file species map written in in *.smap format.
This specifies the gene counts matrix file. See spimap-prep-duploss for details about this file format and how to generate it.
This argument is optional and can be used to limit the amount of computation performed. Specifically, it specifies an upper bound on the number of gene copies that could occur in any species and family.
This argument is optional and can be used to limit the number of iterations used to find the duplication-loss rates.
This argument is optional and can be used to specify an initial estimate of the duplication rate.
This argument is optional and can be used to specify an initial estimate of the loss rate.
When given, shows this help message.
This program is used to prepare a data set for learning gene and species substitution rate parameters. It takes as input a series of trusted gene trees from one-to-one orthologous gene families. The branch lengths of the gene trees should have units of substitution/site. The gene tree filenames should be given one per line on the standard input of this program.
This specifies the file extension of the input gene tree files.
This specifies the file extension of the input alignment files.
This specifies a known species tree with the file species tree written in newick format.
This specifies which species each gene belongs using the file species map written in in *.smap format.
This specifies the output file for the branch length matrix. This file is a tab delimited matrix. Each row represents one family and each column represents one branch in the species tree. Each cell represents the branch length for that family and species branch.
If given, this specifies that gene trees need to be rooted to match the species tree before processing.
When given, shows this help message.
This specifies a known species tree with the file species tree written in newick format.
This specifies the input branch length matrix. See spimap-prep-rates for details about this file format and how to generate it.
This specifies the output file for writing the substitution rate parameters.
This specifies the maximum number of iterations to run to find the substitution rate parameters.
This optional parameter can be used to specify how many discrete segments to use when approximating the gene rate distribution (default: 20).
This optional parameter can be used to discard outlier trees with very large branch lengths.
This optional parameter can be used to discard branches that have too few substitutions.
When given, shows this help message.
This program implements SPIMAP's gene family model for simulating gene trees and DNA alignments. This program is particular useful for creating data sets for evaluating gene tree reconstruction programs.
This specifies a known species tree with the file species tree written in newick format.
This specifies the substitution rate parameters that should be used for simulation.
This specifies the gene length to simulate.
This specifies the transition/transversion rate to use for sequence simulation.
This specifies the background base frequency to use for sequence simulation.
If given, this specifies the minimum number of genes required for each simulated gene family.
If given, this specifies the maximum number of genes required for each simulated gene family.
This specifies the gene duplication rate to use for simulating each gene tree.
This specifies the gene loss rate to use for simulating each gene tree.
This specifies how many gene trees and alignments to simulate.
This option allows you to start simulating the Nth gene family where N=<starting number>.
If given, species rates are not used.
This specifies the output directory for simulations.
This specifies the output file extension for gene tree files.
This specifies the output file extension for reconciliation files.
This specifies the output file extension for alignment files.
This specifies the output file extension for FASTA files.
This specifies the output file extension for simulation information files.
If given, gene family sub-directories are not created.
If given, the simulation program will try to resume where it left off in simulating gene families.
When given, shows this help message.
Restrictions on gene IDs and species IDs. Due to the file formats that SPIMAP uses, there as several restrictions on what IDs are allowed. Many of these restrictions are common for other similar phylogenetic software. The safest IDs follow these restrictions:
Space characters are discouraged from gene IDs and species IDs since they will probably cause problems with other bioinformatic software that you may use (although SPIMAP can handle spaces). Characters such as parentheses "(" ")" and colons ":" are not allowed because the newick file format uses these characters for describing the structure of the tree.
It is also easier to use gene IDs that have a prefix or suffix that indicates the species ID. For example "human_HOXC5" is a human gene. This is not a requirement, but it does make preparing a gene to species mapping file (*.smap) easier.
SPIMAP uses the FASTA file format for sequences alignments. The file extension is not important and many different extensions are in common use (*.fa, *.mfa, *.fasta, *.align).
Each line starting with ">" indicates a gene name (see below). Note, the entire line after the ">" is used as the gene name. The gene's sequence is given on the following lines and it may be wrapped to any number of columns (or not wrapped at all). Gaps in the alignment are represented with the "-" character.
At this time (version 1.1), SPIMAP can only use DNA sequences. The sequence can be in both upper case and lower case (SPIMAP ignores case) and degeneracy codes can be used ("NnRrYyWwSsKkMmBbDdHhVv"), however at this time SPIMAP treats all degeneracies as completely missing data ("N"). Gaps "-" are also treated as missing data.
>KLLA0C08239g ATGAGTCTCAAACGTGTAGTTGTCACTGGTCTTGGGGCCTACACGCCCCTTGGTTCTACAGTTTCAAAGTCTTGGGCAGG TTTGCTT---GCTGCTAAGCAATCACTAATACCCTTAGATGCTTTCTACAACAGAGAA---GACTTTGCAAAAGTGAAAA AGTTGGTCCCACTAGATACAGCAGTGAGTAGGTTACAT------------------------------------------ >ADL072C ---ATGCATCCCCGAGTGGTCGTGACCGGCATTGGGTGCTATACTCCTCTGGGGCCGTCGCTAGCCCAGTCTTGGAAGGA GCTGTTG---CGAGGGACGAGCGGCCTTGTCAGGCTGCAAGATCTGGCAGAGTACGAGGGCGATTACAAACCACTGTCGA GGCTTATATCCGGTGATCTTCGAGTCGGGAAAGTTGGATTTGAG------------------------------------ >kwal_5828 ---ATGACTTCCAGAGTCGTTGTTACTGGGCTTGGTGCTATCACTCCACTTGGGAGGACTGTTTCCGAGTCATGGAGAGC TTTATTG---GCAGGCAAGTCCGGAATTCGTCCCATTCGCGATCTTCCC------------AATGCTAAAAGCTACGAAG GACACTGTCCTGCATCTGTTGCCGTTGCAGACATTCCTGATTTC------------GATCCA------------------
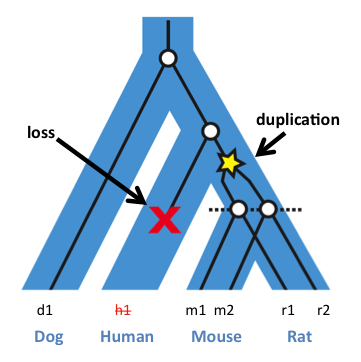
Species trees should be specified using the Newick file format. Beyond the newick format, SPIMAP has only a few additional requirements. First, the species names given in the species tree should match those given in the SMAP file. Second, the branch lengths of the species tree should be expressed in units of time (see figure). Any unit of time can be used (e.g. millions of years, generations, relative units, etc). The only requirement is that the duplication and loss rates are also expressed in compatible units. Therefore, if branch lengths are in millions of years, the duplication rate (specified by spimap's "-D" option) should be in units of duplications/gene/million years.
Naming ancestral nodes. SPIMAP also supports naming ancestral nodes in the species tree using the newick format. For example, the parental node of human and chimp can be named primate using the following syntax:
If ancestral nodes are named, they will be used in the output of the reconciliation mapping.
(((((((scer:7.061760,
spar:7.061760
)n7:4.999680,
smik:12.061440
)n6:5.970600,
sbay:18.032040
)n5:52.682400,
cgla:70.714260
)n4:7.220700,
scas:77.934960
)n3:23.181480,
(
(
agos:78.553260,
klac:78.553260
)n9:10.434960,
kwal:88.988220
)n8:12.128400
)n2:78.883560,
(
(
(
calb:41.275620,
ctro:41.275980
)n12:29.632860,
(
cpar:52.323120,
lelo:52.323120
)n13:18.585720
)n11:31.149540,
(
(
cgui:75.615840,
dhan:75.615840
)n15:14.006880,
clus:89.622720
)n14:12.435660
)n10:77.941620
)n1;

SPIMAP uses a special file format (*.smap) to specify which genes belong to which species. Each line contains two tab-delimited fields:
Only 3 types of gene ID patterns are supported. The pattern can either be an exact matching string, a prefix (denoted "text*"), or a suffix (denoted "*text"). The "*" is the only special wildcard character.
The species ID should be the same as those used in the species tree. All patterns and IDs are case-sensitive.
When mapping a gene ID to a species ID all exact matches are processed first. If no exact match is found, the patterns are then processed in the same order as they appear in the file until a match is found. For example in the SMAP file given below, the gene ID "YALI123" should match the species "ylip", instead of "scer", because the pattern "YALI*" occurs before "Y*".
A* agos orf19* calb CDUG_* cdub CAGL* cgla IPF_* cgla CGUG_* cgui sbay_* sbay scas_* scas smik_* smik spar_* spar SCP* spom YALI* ylip Y* scer Q* scer
When SPIMAP's "-r" option is used, the reconciliation found for the gene tree and species is saved to a file "OUTPUT_PREFIX.recon" (see reconcile example). The reconciliation file format is tab-delimited, where each line has three fields:
Each line specifies the mapping of one node in the gene tree (field 1) to one node or branch in the species tree (field 2). Branches are indicate using the node ID directly below it (i.e. the younger of the two incident nodes). The lines can be given in any order.
If the gene node is a leaf, it will map to a leaf in the species tree and the event field will contain the event "gene". All internal nodes of the gene tree are marked either as speciations (event "spec") or duplications (event "dup"). Specaition nodes map directly to the indicated species node, and duplication nodes map to the indication species branch. The time of the duplication along the species branch is not indicated in this file format nor is it inferred by SPIMAP.
If gene IDs are not given to the ancestral nodes of a gene tree or species tree, SPIMAP will by default name them with "nXXX" where XXX is the preorder traversal of the internal nodes.
KLLA0C08239g klac gene ADL072C agos gene kwal_5828 kwal gene CAGL0J02970g cgla gene scas_g715.48 scas gene smik_6662 smik gene sbay_7039 sbay gene smik_6659 smik gene sbay_7037 sbay gene YER061C scer gene spar_6281 spar gene n10 n5 spec n9 n7 spec n8 n6 spec n7 n5 spec n6 n5 dup n5 n3 spec n4 n3 spec n3 n9 spec n2 n8 spec n1 n2 spec

SPIMAP has several parameters for its substitution rates model. These parameters are learned by the spimap-train-rates program, which saves the parameters in a custom *.params file format (see example below). The spimap program reads these parameters using the "-p" option. Most uses of SPIMAP do not require understanding the contents of a *.params file.
The *.params file format is tab-delimited and each line is processed one at a time.
If the first field of a line is the word "baserate", then the remaining two fields are interpreted as floating point values αG and βG, which are the two parameters, shape and scale, of the inverse-gamma distributed gene-specific rate.
If the first field of the line does not match "baserate", then the first field indicates a species tree branch and the remaining two fields are interpreted as floating point values αi and βi, which are the two parameters, shape and scale, of the gamma distributed species-specific rate. Each branch is indicated by its more recent node. Ancestral nodes are indicated by integers according to preorder traversal.
baserate 6.98457288742 5.98457288742 1 3.28887700831 394.209221588 2 4.64684152603 551.109741211 3 1.13027572632 164.191940308 4 0.610769152641 75.0393371582 5 7.14405012131 927.631103516 6 2.96983885765 238.195861816 7 5.63683271408 632.264831543 8 0.974860072136 94.9837493896 9 0.856632292271 78.6899032593 10 4.64683914185 544.528686523 11 1.92581880093 271.891052246 12 3.84569692612 624.703308105 13 3.14617466927 335.446655273 14 0.699178874493 84.1814575195 15 0.746283352375 137.345901489 scer 8.42576217651 763.305847168 ctro 6.70220327377 999.845153809 scas 9.14448356628 1253.45031738 agos 8.84074497223 801.648925781 sbay 6.95680332184 1048.7590332 kwal 14.3321857452 1962.9083252 dhan 15.7483224869 2699.00878906 smik 10.2562847137 1143.78076172 cgla 9.81903266907 1015.43951416 spar 5.80616807938 799.18963623 calb 8.38038921356 1233.68322754 lelo 9.40990924835 973.772583008 cpar 9.43262672424 1184.28100586 klac 6.6709280014 767.418823242 clus 8.37989234924 881.762878418 cgui 11.9692239761 1187.47314453